神经胶质瘤患者迁移学习的监督白质束分割
IF 11.8
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
在临床设置中,白质束的虚拟解剖代表了监测神经系统状况或支持治疗计划的信息源。通过数据驱动的方法,特别是深度学习模型来执行这项任务,证明了在应用于健康个体时实现高精度的有希望的证据。然而,缺乏大型临床数据集以及健康人群和临床人群之间的巨大差异阻碍了将这些结果转化为患者。在这里,我们首次调查了迁移学习在将健康人群训练的深度学习架构适应胶质瘤患者方面的有效性。重要的是,我们首次提供了域移位及其复杂性的全面表征,将系统(即测量和预处理相关)与肿瘤特异性成分区分开来。我们的研究结果表明:(i)当对患者进行推理时,在大型规范健康人群上训练的模型的性能显著下降;(ii)迁移学习可以成为克服临床数据短缺和管理系统转移的有效策略;(iii)学习模型的微调不能适应肿瘤引起的大的白质变形。结果在五个白质束和三种输入模式测试中是一致的,突出了它们的稳健性和普遍性。我们的工作为推进临床人群的自动白质分割和增强临床转移学习应用提供了有价值的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
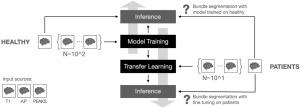
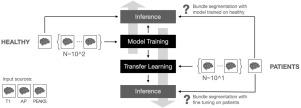
Supervised white matter bundle segmentation in glioma patients with transfer learning
In clinical settings, the virtual dissection of white matter tracts represents an informative source of information for monitoring neurological conditions or to support the planning of a treatment. The implementation of this task through data-driven methodologies and, in particular, deep learning models demonstrates promising evidence of achieving high accuracy when applied to healthy individuals. However, the lack of large clinical datasets and the profound differences between healthy and clinical populations hinder the translation of these results to patients. Here, we investigated for the first time the effectiveness of transfer learning in adapting a deep learning architecture trained on a healthy population to glioma patients. Importantly, we provided the first thorough characterization of domain shift and its complexity, distinguishing systematic (i.e. measurement and pre-processing related) from tumor-specific components. Our results suggest that (i) models trained on a large normative healthy population have a significant performance drop when the inference is carried out on patients; (ii) transfer learning can be an effective strategy to overcome the shortage of clinical data and to manage the systematic shift; (iii) fine-tuning of the learning model cannot accommodate large white matter deformations induced by the tumor. The results were coherent across the five white matter bundles and the three input modalities tested, highlighting their robustness and generalizability. Our work provides valuable insights for advancing automated white matter segmentation in clinical populations and enhancing clinical transfer learning applications.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


