随机流行病模型在线推理的顺序蒙特卡罗平方
IF 2.4
3区 医学
Q2 INFECTIOUS DISEASES
引用次数: 0
摘要
有效的流行病建模和监测需要计算效率高的方法,这些方法可以在获得新数据时不断更新参数估计。本文探讨了序列蒙特卡罗平方(O-SMC2)的在线变体在随机易感-暴露-感染-去除(SEIR)模型中的应用,用于实时流行病跟踪。O-SMC2的优势在于它能够仅利用最近观测的固定窗口,使用粒子Metropolis-Hastings核来更新参数估计。与需要处理所有过去观测值的标准SMC2相比,该功能可以及时更新参数,并显著提高计算效率。首先,我们证明了O-SMC2在真实参数值和观测过程都已知的模拟流行病数据上的效率。然后,我们对来自爱尔兰的真实世界的COVID-19数据集进行了应用,成功地跟踪了疫情,并估计了该疾病的随时间变化的繁殖数量。我们的研究结果表明,O-SMC2提供了准确的在线估计静态和动态流行病学参数,同时大大降低了计算成本。这些发现突出了O-SMC2在实时流行病监测和支持适应性公共卫生干预方面的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
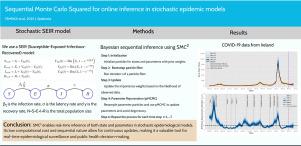
Sequential Monte Carlo Squared for online inference in stochastic epidemic models
Effective epidemic modeling and surveillance require computationally efficient methods that can continuously update parameter estimates as new data becomes available. This paper explores the application of an online variant of Sequential Monte Carlo Squared (O-SMC) to the stochastic Susceptible–Exposed–Infectious–Removed (SEIR) model for real-time epidemic tracking. The advantage of O-SMC lies in its ability to update parameter estimates using a particle Metropolis–Hastings kernel by only utilizing a fixed window of recent observations. This feature enables timely parameter updates and significantly enhances computational efficiency compared to standard SMC, which requires processing all past observations. First, we demonstrate the efficiency of O-SMC on simulated epidemic data, where both the true parameter values and the observation process are known. We then make an application to a real-world COVID-19 dataset from Ireland, successfully tracking the epidemic and estimating a time-dependent reproduction number of the disease. Our results show that O-SMC provides accurate online estimates of both static and dynamic epidemiological parameters while substantially reducing computational cost. These findings highlight the potential of O-SMC for real-time epidemic monitoring and supporting adaptive public health interventions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Epidemics
INFECTIOUS DISEASES-
CiteScore
6.00
自引率
7.90%
发文量
92
审稿时长
140 days
期刊介绍:
Epidemics publishes papers on infectious disease dynamics in the broadest sense. Its scope covers both within-host dynamics of infectious agents and dynamics at the population level, particularly the interaction between the two. Areas of emphasis include: spread, transmission, persistence, implications and population dynamics of infectious diseases; population and public health as well as policy aspects of control and prevention; dynamics at the individual level; interaction with the environment, ecology and evolution of infectious diseases, as well as population genetics of infectious agents.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


