利用生成式机器学习增强低数据体制下的神经分子成像分类:酒精使用障碍的HDAC PET/MR成像案例研究
引用次数: 0
摘要
正电子发射断层扫描(PET)是研究脑相关疾病的重要方式。然而,数据稀缺,尤其是神经表观遗传酶等新分子靶点的数据稀缺,加上难以招募的患者群体,限制了机器学习(ML)模型的发展。我们的主要目标是加强神经分子成像数据的单学科分类,促进生物标志物的发现。我们在酒精使用障碍(AUD)中使用组蛋白去乙酰化酶(HDAC) PET/MR成像证明了我们的方法。方法我们提出了催化训练管道,这是一个利用Wasserstein条件生成对抗网络(WCGAN)生成的高质量合成数据增强真实成像数据的框架。使用[11C]Martinostat PET/MR成像,我们提取了代表8个扣带亚区HDAC酶表达密度的1-D标准化摄取值比(SUVR)表格特征。这些被用来训练和测试ML分类器,包括支持向量机(SVM)、XGBoost和随机森林,在留一交叉验证下。结果在训练过程中集成合成数据显著提高了分类准确率:XGBoost和Random Forest的分类准确率为+26%(从59%提高到85%),SVM的分类准确率为+18%(从70%提高到88%)。合成样本提高了模型的泛化能力。关键的半球和分区域扣带HDAC模式也被确定为潜在的生物标志物。我们的研究结果表明,生成式人工智能可以帮助克服低数据机制神经成像应用中的数据稀缺性。Catalysis Training提供了一种可扩展的策略,以增强机器学习驱动的生物标志物发现和疾病分类,特别是对于罕见或难以研究的疾病,如AUD。在临床上,[11C]Martinostat PET/MR测量的扣带HDAC表达有望作为AUD的客观生物标志物,补充基于dsm的诊断并为新的治疗策略提供信息。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Enhancing neuromolecular imaging classification in low-data regimes with generative machine learning: A case study in HDAC PET/MR imaging of alcohol use disorder
Introduction
Positron Emission Tomography (PET) is a vital modality for investigating brain related disorders. However, data scarcity especially for novel molecular targets like neuroepigenetic enzymes combined with difficult-to-recruit patient populations limits the development of machine learning (ML) models. Our primary objective is to enhance single-subject classification of neuromolecular imaging data and facilitate biomarker discovery. We demonstrate our approach using histone deacetylase (HDAC) PET/MR imaging in Alcohol Use Disorder (AUD).
Methods
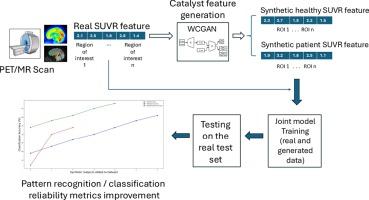
We propose Catalysis Training pipeline, a framework that augments real imaging data with high-quality synthetic data generated by a Wasserstein Conditional Generative Adversarial Network (WCGAN). Using [11C]Martinostat PET/MR imaging, we extracted 1-D standardized uptake value ratio (SUVR) tabular features representing HDAC enzyme expression density across eight cingulate subregions. These were used to train and test ML classifiers, including Support Vector Machine (SVM), XGBoost, and Random Forest, under leave-one-out cross-validation.
Results
Integrating synthetic data in the training process improved classification accuracy significantly: +26% for XGBoost and Random Forest (from 59% to 85%), and +18% for SVM (from 70% to 88%). Synthetic samples improved model generalizability. Key hemispheric and subregional cingulate HDAC patterns were also identified as potential biomarkers.
Conclusion
Our results demonstrate that generative AI can help overcome data scarcity in low-data regime neuroimaging applications. Catalysis Training provides a scalable strategy to enhance ML-driven biomarker discovery and disease classification, especially for rare or difficult-to-study disorders like AUD. Clinically, cingulate HDAC expression measured by [11C]Martinostat PET/MR shows promise as an objective biomarker for AUD, complementing DSM-based diagnosis and informing novel treatment strategies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Neuroscience informatics
Surgery, Radiology and Imaging, Information Systems, Neurology, Artificial Intelligence, Computer Science Applications, Signal Processing, Critical Care and Intensive Care Medicine, Health Informatics, Clinical Neurology, Pathology and Medical Technology
自引率
0.00%
发文量
0
审稿时长
57 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


