治疗性假单胞菌噬菌体Pa223的高分辨率冷冻电镜分析。
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
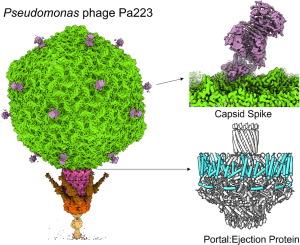
低温电子显微镜(cryo-EM)分析噬菌体是破译病毒组成和构象可塑性的一种有价值的方法。在这项研究中,我们展示了假单胞菌病毒Pa223的高分辨率结构图谱,这是一种来自Bruynoghevirus属的噬菌体,最近被用于临床治疗囊性纤维化和非囊性纤维化支气管扩张,以及同情护理。通过结合生物信息学、蛋白质组学、低温电镜单粒子分析和局部重建,我们对形成二十面体衣壳和不可收缩尾部的8条结构多肽链进行了注释和原子模型的构建。我们发现Pa223衣壳由一个具有独特三β螺旋折叠的刺突蛋白装饰,该蛋白在数据库中没有结构同源物。Pa223尾部具有6条向上延伸的三聚体尾纤维,与噬菌体T7相似,但比它们短。与T7不同的是,Pa223的尾部由两个首尾相连的适配器延伸,并由三聚体尾针密封,类似于p22样噬菌体。我们发现了一种结合在门静脉蛋白外周的蛋白,定位类似于射蛋白gp72,该蛋白在假单胞菌噬菌体DEV中被发现,DEV是一种Litunavirus噬菌体,也是重新分类的血吸虫病毒科成员。根据这一结构线索,我们鉴定出了Pa223喷射蛋白gp53、gp54和gp56,它们在生物信息学上与噬菌体T7的相似程度高于血吸虫病毒科。因此,Pa223含有与p22样、t7样和Litunavirus噬菌体相似的多种结构元件,为了解bruynoghevirus中喷射蛋白的进化提供了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
High-resolution Cryo-EM Analysis of the Therapeutic Pseudomonas Phage Pa223
Cryogenic electron microscopy (cryo-EM) analysis of bacteriophages is a valuable method for deciphering virus composition and conformational plasticity. In this study, we present a high-resolution structural atlas of the Pseudomonas virus Pa223, a phage from the Bruynoghevirus genus that has recently been used in clinical cocktails for treating cystic fibrosis and non-cystic fibrosis bronchiectasis, as well as for compassionate care. By combining bioinformatics, proteomics, cryo-EM single particle analysis, and localized reconstruction, we annotated and built atomic models for eight structural polypeptide chains that form the icosahedral capsid and noncontractile tail. We discovered that the Pa223 capsid is decorated by a spike protein with a unique triple-β helix fold that has no structural homologs in the database. The Pa223 tail features six trimeric tail fibers extending upward, similar to but shorter than those found in phage T7. Unlike T7, the Pa223 tail is extended by two head-to-tail adaptors and sealed by a trimeric tail needle, similar to P22-like phages. We identified a protein bound around the outer perimeter of the portal protein, positioned similarly to the ejection protein gp72, which was identified in the Pseudomonas phage DEV, a Litunavirus phage, and a member of the reclassified Schitoviridae family. This structural clue led us to identify the Pa223 ejection proteins gp53, gp54, and gp56, which bioinformatically resemble those of phage T7 more closely than Schitoviridae. Thus, Pa223 contains various structural elements similar to those in P22-like, T7-like, and Litunavirus phages, providing a foundation for understanding the evolution of ejection proteins in Bruynogheviruses.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


