用于创建和评估药物错误合成数据集的人工智能驱动方法。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:建立完整的用药差错(ME)数据集。这将有助于解决在医疗保健应用程序中开发机器学习模型时对真实世界数据访问受限的挑战。方法:我们使用基于变压器的模型(GPT-4、LLAMA3和Mistral)创建法语合成数据集。这些模型生成了捕获ME类型可变性的各种描述。我们通过医疗保健专业人员的专家评估和人工智能驱动的分析来评估合成数据集的有效性,以测试其现实性及其在训练机器学习模型用于ME分类中的实用性。结果:合成数据集在表示不同ME场景方面具有较高的准确性和真实感。专家评估确认数据集与现实世界的ME数据相似。人工智能驱动的评估还表明,在合成数据上训练的模型取得了稳健的分类性能,验证了数据集在开发有效的ME分类工具方面的实用性。结论:所提出的方法证明了大型语言模型在生成真实的法语综合ME报告方面的潜力。在200份评估报告中,70%的零注射输出被认为低于预期,而80%的单注射和少注射输出被临床专家认为有效或经过轻微修改后有效。此外,在800个合成报告上训练的分类器在对真实数据进行测试时获得了高达0.78的f1分数。这些结果证实,在现实世界数据有限或不可用的情况下,合成数据可以有效地支持人工智能驱动的ME分析。本文章由计算机程序翻译,如有差异,请以英文原文为准。
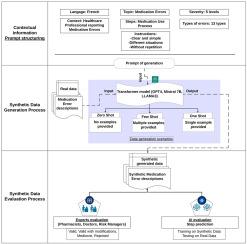
AI-driven approach for creating and evaluating a synthetic dataset for Medication Errors
Objective:
This study aims to create a complete Medication Error (ME) dataset. This will help to address the challenge of limited access to real-world data for developing machine learning models in healthcare applications.
Methods:
We use transformer-based models (GPT-4, LLAMA3, and Mistral) to create our synthetic dataset in French. These models generate a diverse range of descriptions that capture the variability of ME types. We assess the effectiveness of our synthetic dataset through expert evaluations by healthcare professionals and an AI-driven analysis, to test its realism and its utility in training machine learning models for ME classification.
Results:
The synthetic dataset demonstrates high accuracy and realism in representing diverse ME scenarios. Expert evaluation confirms that the dataset is similar to real-world ME data. The AI-driven evaluation also shows that models trained on synthetic data achieved robust classification performance, validating the dataset’s utility for the development of effective ME classification tools.
Conclusion:
The proposed approach demonstrates the potential of large language models to generate realistic synthetic ME reports in French. Out of 200 evaluated reports, 70% of zero-shot outputs were deemed below expectations, while 80% of one-shot and few-shot outputs were considered valid or valid with minor revisions by clinical experts. Furthermore, classifiers trained on 800 synthetic reports attained an F1-score of up to 0.78 when tested on real data. These results confirm that synthetic data can effectively support AI-driven ME analysis in contexts where real-world data is limited or unavailable.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


