单细胞RNA测序揭示了肺腺癌中棕榈酰化驱动的细胞异质性和预后生物标志物
IF 5
2区 医学
Q2 Medicine
引用次数: 0
摘要
肺腺癌(LUAD)以肿瘤细胞内的显著变异为特征,并且仍然是全球癌症死亡的主要原因。棕榈酰化是一种动态的脂质修饰,发生在蛋白质合成之后,影响各种癌症相关蛋白的行为和寿命。然而,其在LUAD患者细胞复杂性形成和预后预测中的作用尚未完全明确。方法我们检测了LUAD样本的单细胞RNA测序数据集,以鉴定不同的恶性细胞群。使用GSVA和ssGSEA技术估计棕榈酰化相关基因的活性。为了进一步定义细胞特征,我们应用了拷贝数变异图谱、伪时间进展模型、转录因子调控评分和细胞-细胞相互作用分析。利用棕榈酰化模式最突出的集群(C1)中的标记基因建立了一个12基因风险模型。该模型在癌症基因组图谱(TCGA)数据集上进行训练,并使用单独的GEO数据集进行验证。为了评估肿瘤免疫环境,我们分析了不同风险水平下免疫细胞的存在和肿瘤突变负担。在LUAD细胞培养中进行了天冬氨酸β -羟化酶(ASPH)上调和沉默的实验室实验,以验证其生物学意义。结果我们鉴定了6个肿瘤细胞亚群(C0至C5),其中C1簇显示棕榈酰化水平峰值,明显的基因组改变,与基质细胞和免疫细胞的通讯更强。12基因模型有效地将LUAD患者分为高风险和低风险,显示出显著的生存差异(p <;0.001),并且在时间相关的ROC分析中表现良好。高危组患者肿瘤突变负担加重,肿瘤环境免疫抑制性增强。实验室结果显示,提高ASPH表达可促进细胞生长、运动和上皮-间质转化。相反,降低ASPH水平会引发细胞死亡并降低侵袭性。结论针对棕榈酰化的sour单细胞分析揭示了LUAD中肿瘤多样性的新维度,并建立了经过验证的12个基因风险特征。功能研究强调了ASPH作为治疗靶标的有希望的候选者。这些结果加深了我们对棕榈酰化相关途径的理解,并为LUAD的预后预测和基于精度的治疗策略提供了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
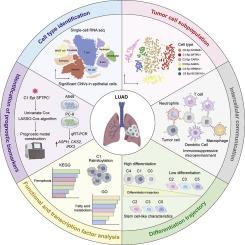
Single-cell RNA sequencing reveals palmitoylation-driven cellular heterogeneity and prognostic biomarkers in lung adenocarcinoma
Background
Lung adenocarcinoma (LUAD) is marked by significant variation within tumor cells and continues to be a major global cause of cancer deaths. Palmitoylation is a dynamic lipid-based modification that occurs after protein synthesis and influences the behavior and lifespan of various cancer-related proteins. However, its role in shaping cellular complexity and predicting outcomes in LUAD patients is not yet fully clarified.
Methods
We examined single-cell RNA sequencing datasets from LUAD samples to identify distinct malignant cell groups. Palmitoylation-related gene activity was estimated using GSVA and ssGSEA techniques. To further define cellular characteristics, we applied copy number variation mapping, pseudotime progression modeling, transcription factor regulatory scoring, and cell–cell interaction analyses. A 12-gene risk model was developed using marker genes from the cluster (C1) with the most prominent palmitoylation pattern. This model was trained on The Cancer Genome Atlas (TCGA) dataset and confirmed using separate GEO datasets. To evaluate tumor immune context, we analyzed immune cell presence and tumor mutational burden across different risk levels. Laboratory experiments involving both upregulation and silencing of aspartate beta-hydroxylase (ASPH) in LUAD cell cultures were conducted to validate its biological significance.
Results
We identified six tumor cell subsets (C0 to C5), with cluster C1 showing peak palmitoylation levels, distinct genomic alterations, and stronger communication with stromal and immune cells. The 12-gene model effectively categorized LUAD patients into high- and low-risk profiles, showing marked survival differences (p < 0.001) and strong performance in time-dependent ROC analysis. Patients in the high-risk group had increased tumor mutational burden and a more immunosuppressive tumor environment. Laboratory findings revealed that raising ASPH expression promoted cell growth, motility, and epithelial–mesenchymal transition. In contrast, reducing ASPH levels triggered cell death and decreased invasiveness.
Conclusions
Our single-cell analysis focused on palmitoylation reveals new dimensions of tumor diversity in LUAD and establishes a validated 12-gene risk signature. Functional studies highlight ASPH as a promising candidate for therapeutic targeting. These results deepen our understanding of palmitoylation-associated pathways and present a foundation for both outcome prediction and precision-based treatment strategies in LUAD.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Translational Oncology
ONCOLOGY-
CiteScore
8.40
自引率
2.00%
发文量
314
审稿时长
54 days
期刊介绍:
Translational Oncology publishes the results of novel research investigations which bridge the laboratory and clinical settings including risk assessment, cellular and molecular characterization, prevention, detection, diagnosis and treatment of human cancers with the overall goal of improving the clinical care of oncology patients. Translational Oncology will publish laboratory studies of novel therapeutic interventions as well as clinical trials which evaluate new treatment paradigms for cancer. Peer reviewed manuscript types include Original Reports, Reviews and Editorials.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


