宏基因组学在水生生态系统中的潜在应用及未来展望。
IF 2.4
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
摘要
宏基因组学在促进我们对各种生态系统中微生物群落及其功能贡献的理解方面起着至关重要的作用。通过直接对环境样本(如土壤、水、空气和人体)的DNA进行测序,宏基因组学能够识别以前不可培养或未知的微生物,为其生态功能提供关键见解。除了分类分类,宏基因组分析还揭示了功能基因和代谢途径,促进了酶、生物活性化合物和其他分子在农业、生物技术和医学中的应用。本文综述了宏基因组学在环境监测中的广泛应用,包括样品采集、高通量测序、数据分析和解释。我们回顾了不同的测序平台、文库制备方法以及用于质量控制、序列组装以及分类和功能注释的先进生物信息学工具。特别关注宏基因组学在评估微生物对环境胁迫、污染物降解、疾病出现和气候变化的反应方面的作用。还审查了微生物生物指标在水生生态系统监测和毒理学评估中的应用。对当前生物信息学管道在处理大规模宏基因组数据集方面的有效性进行了全面评估。随着全球环境压力的加剧,综合元组学方法,包括全基因组元组学,将对理解自然和受影响生态系统中微生物组的复杂性、功能和动态至关重要。本文章由计算机程序翻译,如有差异,请以英文原文为准。
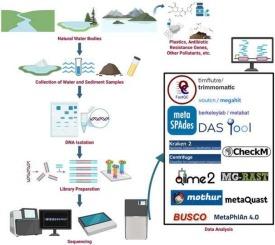
Potential applications and future prospects of metagenomics in aquatic ecosystems
Metagenomics plays a vital role in advancing our understanding of microbial communities and their functional contributions in various ecosystems. By directly sequencing DNA from environmental samples such as soil, water, air, and the human body. Metagenomics enables the identification of previously uncultivable or unknown microorganisms, offering key insights into their ecological functions. Beyond taxonomic classification, metagenomic analyses reveal functional genes and metabolic pathways, facilitating the discovery of enzymes, bioactive compounds, and other molecules with applications in agriculture, biotechnology, and medicine. This review discusses the broad applications of metagenomics in environmental monitoring, encompassing sample collection, high-throughput sequencing, data analysis and interpretation. We review different sequencing platforms, library preparation methods, and advanced bioinformatics tools used for quality control, sequence assembly, and both taxonomic and functional annotation. Special focus is given to the role of metagenomics in evaluating microbial responses to environmental stress, contaminant degradation, disease emergence, and climate change. The use of microbial bioindicators for aquatic ecosystem monitoring and toxicological assessments is also examined. A comprehensive evaluation of current bioinformatics pipelines is provided for their effectiveness in processing large-scale metagenomic datasets. As global environmental pressures intensify, integrative meta-omics approaches, including whole-genome metagenomics, will become crucial for understanding the complexity, functions, and dynamics of microbiomes in both natural and affected ecosystems.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Gene
生物-遗传学
CiteScore
6.10
自引率
2.90%
发文量
718
审稿时长
42 days
期刊介绍:
Gene publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


