客户蛋白的细微变化决定了细菌对Hsp90的依赖性。
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
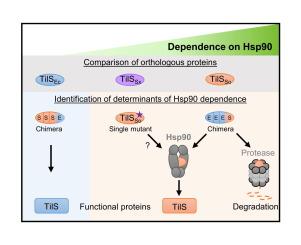
伴侣蛋白确保蛋白质稳态,并在物种间保持保守。依赖atp的伴侣蛋白Hsp90存在于细菌和真核生物中,在那里它稳定和激活广泛的被称为客户的底物蛋白。然而,是什么决定了一种蛋白质是否依赖于Hsp90仍然是一个悬而未决的问题。在这里,我们重点研究了希瓦氏菌中的细菌伴侣Hsp90及其专性客户端TilS(称为TilSSo)。尽管热应激条件下,Hsp90通过保护必需蛋白tilso不被蛋白酶hslv降解而在s.o idensis中不可或缺,但Hsp90在大肠杆菌中是不可或缺的,这表明大肠杆菌TilS (TilSEc)是独立于Hsp90的。因此,我们比较了TilS同源物的体外稳定性、体内降解和与Hsp90的相互作用,以确定Hsp90依赖性的决定因素。我们发现,与TilSSo相比,TilSEc更稳定,在没有Hsp90的情况下不被蛋白酶降解,也不与Hsp90相互作用,表明TilSEc不是Hsp90的客户端。TilSSo和TilSEc之间的嵌合以及定向诱变揭示了TilSSo的一个区域,该区域是蛋白酶降解和Hsp90保护的关键。与这些结果一致的是,在热胁迫下,产生TilSEc的金丝桃生长不再依赖于Hsp90。相反,Hsp90对产生tilso而不是TilSEc的大肠杆菌的生长至关重要。总之,我们的工作揭示了蛋白质特异性特征,如稳定性和降解敏感性,可以确定体内的同源蛋白是否需要细菌Hsp90伴侣蛋白。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Subtle Variations in a Client Protein Determine Bacterial Hsp90 Dependence
Chaperones ensure protein homeostasis and are conserved across species. The ATP-dependent chaperone Hsp90 is present from bacteria to eukaryotes, where it stabilizes and activates a wide range of substrate proteins called clients. However, what determines whether a protein depends on Hsp90 remains an open question. Here, we focused on the bacterial chaperone Hsp90 and its obligate client TilS (referred to as TilSSo) in the bacterium Shewanella oneidensis. Although Hsp90 is indispensable in S. oneidensis under heat stress by protecting the essential protein TilSSo from degradation by the protease HslUV, Hsp90 is dispensable in Escherichia coli, suggesting that E. coli TilS (TilSEc) is Hsp90 independent. We therefore compared the TilS orthologs with respect to in vitro stability, in vivo degradation, and interaction with Hsp90 to identify determinants of Hsp90 dependence. We found that in contrast to TilSSo, TilSEc was more stable, was not degraded by protease in the absence of Hsp90, and did not interact with Hsp90, indicating that TilSEc is not a client of Hsp90. Chimeras between TilSSo and TilSEc as well as directed mutagenesis revealed a region of TilSSo that is key for protease degradation and Hsp90 protection. Consistent with these results, the growth of S. oneidensis producing TilSEc was no longer dependent on Hsp90 under heat stress. Conversely, Hsp90 became essential for the growth of E. coli that produced TilSSo instead of TilSEc. Altogether, our work reveals that protein-specific features, such as stability and degradation sensitivity, can determine whether orthologous proteins require the bacterial Hsp90 chaperone in vivo.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


