空间定量代谢组学能够识别中风后远端和持续的同侧皮质代谢重编程
IF 20.8
1区 医学
Q1 ENDOCRINOLOGY & METABOLISM
引用次数: 0
摘要
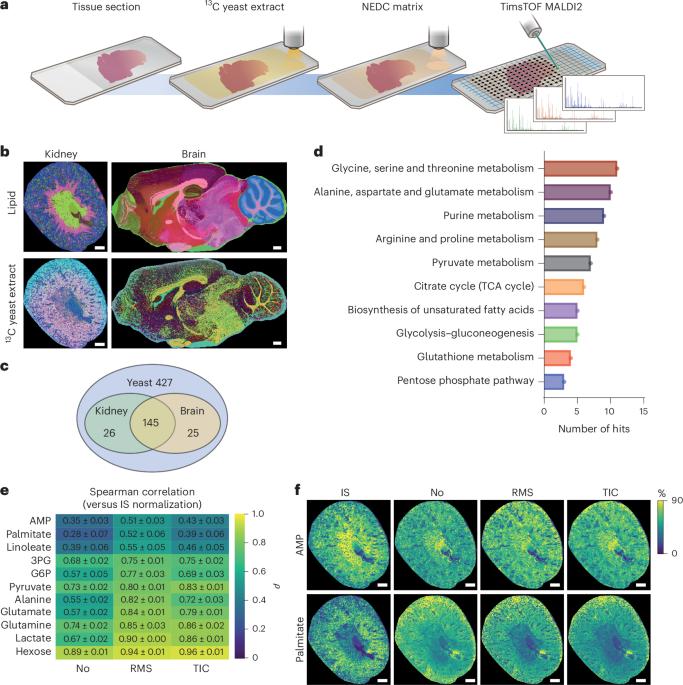
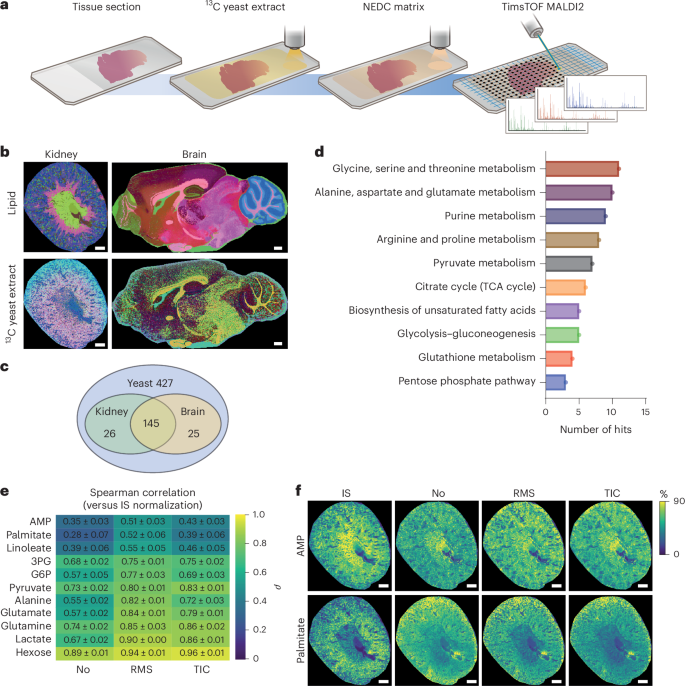
质谱成像(MSI)已成为空间生物学研究的基石。然而,该技术固有的各种因素限制了基于msi的空间代谢组学的定量能力,从而限制了可靠的解释。在这里,我们开发了一种改进的定量MSI工作流程,基于同位素13c标记的酵母提取物作为内部标准,以克服这些缺陷。使用大脑和肾脏组织,我们证明这种方法可以量化200多种代谢特征。将我们的工作流程应用到卒中模型中,不仅可以绘制梗死区和梗死周围区域随时间的代谢重塑图谱,还可以在组织学未受影响的同侧感觉运动皮层中发现迄今为止未被注意到的远程代谢重塑。在中风后第7天,与对侧皮质相比,发现神经保护性赖氨酸水平升高,兴奋性谷氨酸水平降低。到中风后第28天,赖氨酸和谷氨酸水平恢复正常,而尿苷二磷酸n -乙酰氨基葡萄糖和亚油酸的前体池持续减少,这与脆弱性有关。重要的是,不使用内部标准的传统规范化策略无法可视化这些差异。使用13c标记的酵母提取物作为标准化策略,在定量的基于msi的空间代谢组学中建立了一个范例,大大提高了可靠性和解释强度。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Spatial quantitative metabolomics enables identification of remote and sustained ipsilateral cortical metabolic reprogramming after stroke
Mass spectrometry imaging (MSI) has become a cornerstone of spatial biology research. However, various factors that are intrinsic to the technology limit the quantitative capacity of MSI-based spatial metabolomics and thus reliable interpretation. Here we developed an improved quantitative MSI workflow, based on isotopically 13C-labelled yeast extract as internal standards, to overcome these pitfalls. Using brain and kidney tissue, we demonstrate that this approach allows for quantification of more than 200 metabolic features. Applying our workflow to a stroke model allowed us to not only map metabolic remodelling of the infarct and peri-infarct area over time, but also discover hitherto unnoted remote metabolic remodelling in the histologically unaffected ipsilateral sensorimotor cortex. At day 7 post-stroke, increased levels of neuroprotective lysine and reduced excitatory glutamate levels were found when compared with the contralateral cortex. By day 28 post-stroke, lysine and glutamate levels appeared normal, while decreased precursor pools of uridine diphosphate N-acetylglucosamine and linoleate persisted that were previously linked to vulnerability. Importantly, traditional normalization strategies not using internal standards were unable to visualize these differences. Using 13C-labelled yeast extracts as a normalization strategy establishes a paradigm in quantitative MSI-based spatial metabolomics that greatly enhances reliability and interpretive strength. Wang et al. report a workflow to perform quantitative mass spectrometry imaging and provide insight into spatial metabolic remodelling undergone in the mouse brain upon ischaemia.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature metabolism
ENDOCRINOLOGY & METABOLISM-
CiteScore
27.50
自引率
2.40%
发文量
170
期刊介绍:
Nature Metabolism is a peer-reviewed scientific journal that covers a broad range of topics in metabolism research. It aims to advance the understanding of metabolic and homeostatic processes at a cellular and physiological level. The journal publishes research from various fields, including fundamental cell biology, basic biomedical and translational research, and integrative physiology. It focuses on how cellular metabolism affects cellular function, the physiology and homeostasis of organs and tissues, and the regulation of organismal energy homeostasis. It also investigates the molecular pathophysiology of metabolic diseases such as diabetes and obesity, as well as their treatment. Nature Metabolism follows the standards of other Nature-branded journals, with a dedicated team of professional editors, rigorous peer-review process, high standards of copy-editing and production, swift publication, and editorial independence. The journal has a high impact factor, has a certain influence in the international area, and is deeply concerned and cited by the majority of scholars.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


