与细菌转录因子SqrR/YgaV结合的血红素催化硫化氢向多硫化物的氧依赖性转化,以调节基因表达
IF 11.9
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
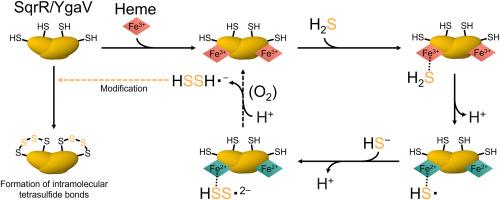
硫化氢(H2S)和多硫化物是细菌中重要的信号分子,在调节氧化应激和氧化还原平衡中发挥着独特的作用。本研究探讨了荚膜红杆菌(Rhodobacter capsulatus)的SqrR和大肠杆菌(Escherichia coli)的YgaV两种同源转录因子硫感知和调控功能的分子机制。体外硫醇特异性标记和SDS-PAGE分析表明,在好氧和厌氧条件下,载脂蛋白sqrr和载脂蛋白ygav对多硫化物有选择性和敏感的反应,而不是对H2S本身。紫外可见光谱表明,血红素的配位状态随半胱氨酸氧化还原状态的变化而变化:还原半胱氨酸支持六坐标血红素,而氧化成四硫交联导致五坐标血红素。重要的是,血红素结合在有氧条件下增强H2S对半胱氨酸的氧化,而在厌氧条件下则没有,这表明氧气促进了血红素介导的多硫化物的生成和利用。相反,血红素结合抑制两种蛋白质在厌氧条件下对多硫化物的半胱氨酸反应性,这种抑制由血红素铁的氧化还原状态调节。这些发现表明,血红素结合通过在好氧条件下促进H2S氧化半胱氨酸和在厌氧条件下抑制多硫化物的反应性来调节硫的反应性。这项工作揭示了细菌转录因子整合氧化还原线索和硫代谢的环境依赖调节机制,揭示了它们对波动氧和硫环境的进化适应。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Heme bound to the bacterial transcription factor SqrR/YgaV catalyzes oxygen-dependent conversion of hydrogen sulfide to polysulfide for regulated gene expression
Hydrogen sulfide (H2S) and polysulfide are critical signaling molecules in bacteria, with distinct roles in regulating oxidative stress and redox balance. This study investigates the molecular mechanisms underlying the sulfur sensing and regulatory functions of two homologous transcription factors, SqrR from Rhodobacter capsulatus and YgaV from Escherichia coli. In vitro thiol-specific labeling and SDS–PAGE analyses demonstrate that apo-SqrR and apo-YgaV respond selectively and sensitively to polysulfides, rather than H2S itself, under both aerobic and anaerobic conditions. UV–visible spectroscopy demonstrated that the coordination state of the heme changes depending on the cysteine redox status: reduced cysteines support a six-coordinate heme, while oxidation to tetrasulfide crosslink leads to a five-coordinate state. Importantly, heme binding enhances cysteine oxidation by H2S under aerobic conditions, but not under anaerobic conditions, indicating that oxygen facilitates heme-mediated generation and utilization of polysulfides. In contrast, heme binding suppresses cysteine reactivity toward polysulfides under anaerobic conditions in both proteins, with this suppression modulated by the redox state of the heme iron. These findings suggest that heme binding regulates sulfur responsiveness by promoting cysteine oxidation by H2S under aerobic conditions and suppressing polysulfide reactivity under anaerobic conditions. This work reveals a context-dependent regulatory mechanism by which bacterial transcription factors integrate redox cues and sulfur metabolism, shedding light on their evolutionary adaptation to fluctuating oxygen and sulfur environments.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Redox Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-
CiteScore
19.90
自引率
3.50%
发文量
318
审稿时长
25 days
期刊介绍:
Redox Biology is the official journal of the Society for Redox Biology and Medicine and the Society for Free Radical Research-Europe. It is also affiliated with the International Society for Free Radical Research (SFRRI). This journal serves as a platform for publishing pioneering research, innovative methods, and comprehensive review articles in the field of redox biology, encompassing both health and disease.
Redox Biology welcomes various forms of contributions, including research articles (short or full communications), methods, mini-reviews, and commentaries. Through its diverse range of published content, Redox Biology aims to foster advancements and insights in the understanding of redox biology and its implications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


