CLEAR:通过持续学习支持临床证据生命周期的愿景。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
人类对疾病、治疗和预防技术的知识在不断发展。使用人类受试者的随机对照试验产生临床证据的过程明显缓慢且效率低下。学习健康系统(LHS)的提出是为了通过知识、实践和数据的循环促进个人和人群健康的持续改善。然而,对支持临床决策的高质量证据的需求与现有证据之间的差距继续扩大。虽然目前的LHS愿景明确了现实世界数据(RWD)的整合,但RWD的快速生成往往超过了有效证据合成和实施的速度。考虑到这一点,我们提出了一个新的框架,通过持续的学习机制,更有效地利用RWD来支持整个临床证据生命周期。该框架由现代数据科学和信息学提供支持,提供了增强的可扩展性和效率。在这一愿景中,RWD通过四个封闭的反馈回路被整合到临床证据生命周期中:1)指导研究优先级和研究设计,2)促进临床指南制定,3)协助指南评估,4)支持共同决策。我们的框架能够快速响应新出现的健康数据和不断变化的医疗保健需求,及时制定临床指南以优化临床建议,并持续改善临床实践和患者结果。这一愿景要求信息学支持高效、可扩展和利益相关者意识的临床证据生命周期。本文章由计算机程序翻译,如有差异,请以英文原文为准。
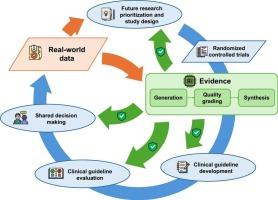
CLEAR: A vision to support clinical evidence lifecycle with continuous learning
Human knowledge of diseases, treatments, and prevention techniques is constantly evolving. The generation of clinical evidence using randomized controlled trials on human subjects occurs notably slowly and inefficiently. The Learning Health System (LHS) has been proposed to facilitate the continuous improvement of individual and population health through a cycle of knowledge, practice, and data. However, the gap between the demand for high-quality evidence to support clinical decisions and the available evidence continues to enlarge. While the current LHS vision articulates the integration of Real-World Data (RWD), the rapid generation of RWD often outpaces the rate of effective evidence synthesis and implementation. Considering this, we propose a new framework that more effectively leverages RWD to support the entire clinical evidence lifecycle through a continuous learning mechanism. This framework, powered by modern data science and informatics, offers enhanced scalability and efficiency. In this vision, specifically, RWD is integrated into the clinical evidence lifecycle via four closed feedback loops: 1) guiding research prioritization and study design, 2) facilitating clinical guideline development, 3) assisting guideline evaluation, and 4) supporting shared decision-making. Our framework enables rapid responsiveness to emerging health data and evolving healthcare needs, timely development of clinical guidelines to optimize clinical recommendations, and sustained improvements in clinical practice and patient outcomes. This vision calls for informatics support for an efficient, scalable, and stakeholder-aware clinical evidence lifecycle.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


