鲁棒T-Loss用于医学图像分割
IF 11.8
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
本文介绍了一种新的用于医学图像分割的鲁棒损失函数T-Loss。T-Loss来源于Student-t分布的负对数似然,并且擅长通过单个参数动态控制其灵敏度来处理噪声掩模。该参数在反向传播过程中进行了优化,从而避免了对噪声标签的范围和分布的额外计算或先验知识的需要。我们在训练过程中对该参数的行为进行了深入分析,揭示了其自适应特性及其在防止噪声记忆中的作用。我们的大量实验表明,就两个公共医疗数据集的骰子分数而言,T-Loss显着优于传统的损失函数,特别是对于皮肤病变和肺分割。此外,T-Loss对模拟人类标注错误的各种类型的模拟标签噪声表现出显著的弹性。我们的研究结果提供了强有力的证据,证明T-Loss是医学图像分割的一种有希望的替代方法,其中数据集中的高水平噪声或异常值是实践中的典型现象。项目网站,包括代码和其他资源,可以在https://robust-tloss.github.io/上找到。本文章由计算机程序翻译,如有差异,请以英文原文为准。
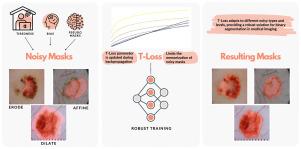
Robust T-Loss for medical image segmentation
This work introduces T-Loss, a novel and robust loss function for medical image segmentation. T-Loss is derived from the negative log-likelihood of the Student-t distribution and excels at handling noisy masks by dynamically controlling its sensitivity through a single parameter. This parameter is optimized during the backpropagation process, obviating the need for additional computations or prior knowledge about the extent and distribution of noisy labels. We provide in-depth analysis of this parameter behavior during training and revealing its adaptive nature and its role in preventing noisy memorization. Our extensive experiments demonstrate that T-Loss significantly outperforms traditional loss functions in terms of dice scores on two public medical datasets, specifically for skin lesion and lung segmentation. Moreover, T-Loss exhibits remarkable resilience to various types of simulated label noise, which mimics human annotation errors. Our results provide strong evidence that T-Loss is a promising alternative for medical image segmentation where high levels of noise or outliers in the dataset are a typical phenomenon in practice. The project website, including code and additional resources, can be found at: https://robust-tloss.github.io/.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


