基于PET/CT放射组学预测体积等效食管鳞状细胞癌患者T2/T3分期
IF 4.8
2区 医学
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的建立并外部验证基于PET/ ct的放射学模型,用于预测体积匹配的食管鳞状细胞癌(ESCC)患者肿瘤侵袭深度(≤T2 vs≥T3)。方法对18F-FDG PET图像进行半自动分割,提取PET和CT扫描的放射学特征。用类内相关系数(ICC)评价特征再现性,结果一致(ICC >;0.95)。采用倾向评分匹配(PSM)来控制肿瘤体积和患者人口统计学。使用主成分分析(PCA)进行降维,然后通过LASSO和MRMR进行特征选择。构建了逻辑回归(LR)、线性判别分析(LDA)、支持向量机(SVM)和随机森林(RF)模型。模型的性能在内部和独立的外部队列中进行评估。结果在内部验证中,PET和PET/CT联合放射学模型优于单独CT模型,LDA和LR分类器的ROC曲线下面积(AUC)均大于0.97。在外部验证中,只有基于PET特征的模型保持了良好的预测性能(LR AUC 0.8438,准确率81.25%;LDA AUC 0.8281,准确率87.5%)。基于CT或PET/CT组合特征的模型无法产生有效的结果,默认为单一类别预测。基于PET特征的模型显示出跨数据集的稳定泛化性。结论基于PET特征和LDA或LR分类器的放射组学模型能准确预测体积等效ESCC患者的肿瘤侵袭深度,具有较强的外部泛化能力。CT或联合特征模型在严格的肿瘤体积限制下可能不可靠。本文章由计算机程序翻译,如有差异,请以英文原文为准。
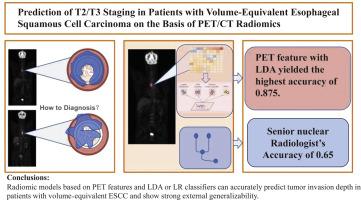
Prediction of T2/T3 Staging in Patients with Volume-Equivalent Esophageal Squamous Cell Carcinoma on the Basis of PET/CT Radiomics
Objectives
To develop and externally validate PET/CT-based radiomic models for predicting tumor invasion depth (≤T2 vs. ≥T3) in patients with esophageal squamous cell carcinoma (ESCC) with volume-matched tumors.
Methods
Semiautomatic segmentation was performed on 18F-FDG PET images, and radiomic features were extracted from PET and coregistered CT scans. Feature reproducibility was evaluated with the intraclass correlation coefficient (ICC), which showed excellent agreement (ICC > 0.95). Propensity score matching (PSM) was applied to control for tumor volume and patient demographics. Dimensionality reduction was conducted using principal component analysis (PCA), followed by feature selection via LASSO and MRMR. Logistic regression (LR), linear discriminant analysis (LDA), support vector machine (SVM), and random forest (RF) models were constructed. Model performance was assessed on internal and independent external cohorts.
Results
In the internal validation, the PET and combined PET/CT radiomic models outperformed the CT-alone models, with the LDA and LR classifiers achieving area under the ROC curve (AUC) greater than 0.97. In the external validation, only the models based on PET features maintained good predictive performance (LR AUC 0.8438, accuracy 81.25 %; LDA AUC 0.8281, accuracy 87.5 %). Models built on CT or combined PET/CT features failed to produce valid results, defaulting to single-class predictions. PET feature-based models demonstrated stable generalizability across datasets.
Conclusions
Radiomic models based on PET features and LDA or LR classifiers can accurately predict tumor invasion depth in patients with volume-equivalent ESCC and show strong external generalizability. CT or combined feature models may not be reliable under stringent tumor volume constraints.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computer methods and programs in biomedicine
工程技术-工程:生物医学
CiteScore
12.30
自引率
6.60%
发文量
601
审稿时长
135 days
期刊介绍:
To encourage the development of formal computing methods, and their application in biomedical research and medical practice, by illustration of fundamental principles in biomedical informatics research; to stimulate basic research into application software design; to report the state of research of biomedical information processing projects; to report new computer methodologies applied in biomedical areas; the eventual distribution of demonstrable software to avoid duplication of effort; to provide a forum for discussion and improvement of existing software; to optimize contact between national organizations and regional user groups by promoting an international exchange of information on formal methods, standards and software in biomedicine.
Computer Methods and Programs in Biomedicine covers computing methodology and software systems derived from computing science for implementation in all aspects of biomedical research and medical practice. It is designed to serve: biochemists; biologists; geneticists; immunologists; neuroscientists; pharmacologists; toxicologists; clinicians; epidemiologists; psychiatrists; psychologists; cardiologists; chemists; (radio)physicists; computer scientists; programmers and systems analysts; biomedical, clinical, electrical and other engineers; teachers of medical informatics and users of educational software.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


