CD8+和CD4 + T-LGL白血病的转录组学景观显示STAT3和STAT5B激活突变的明显影响
IF 13.4
1区 医学
Q1 HEMATOLOGY
引用次数: 0
摘要
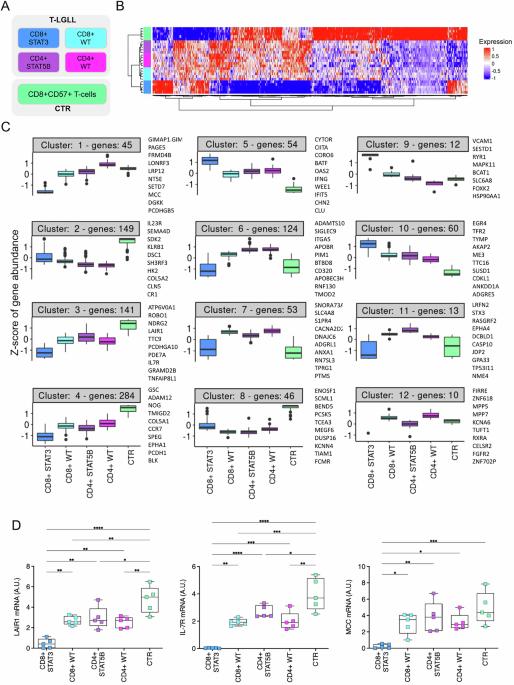
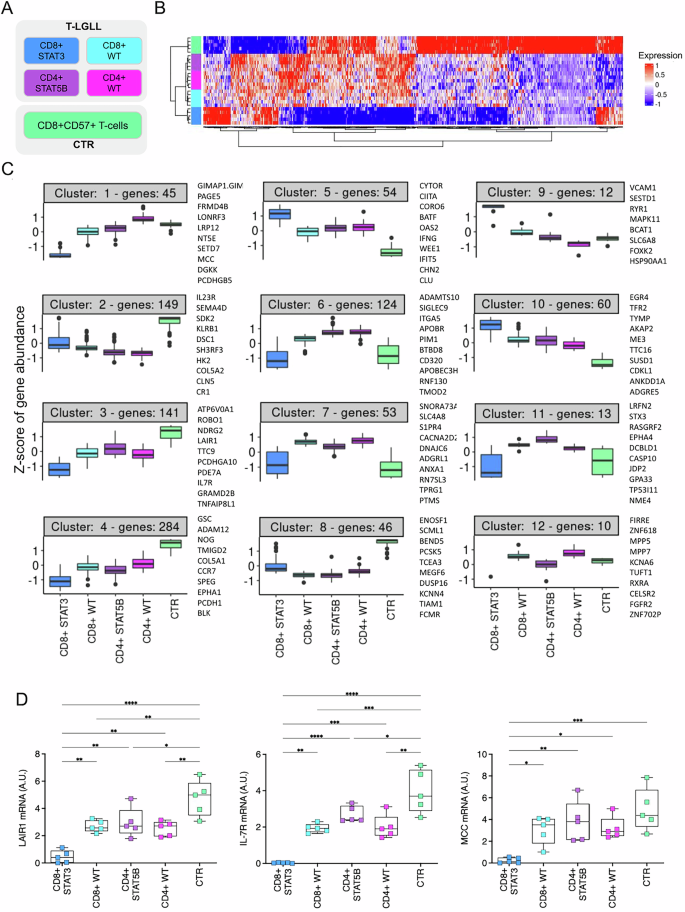
T-LGL白血病(T-LGLL)高临床异质性的生物学基础尚不完全清楚,缺乏有效的治疗方法。通过纯化T-LGLs的rna测序,我们揭示了主要患者亚组的基因表达谱和通路失调,这些亚组由CD8+或CD4+表型和STAT3/STAT5B突变状态定义。总体而言,与对照组相比,T-LGLL患者表现出明显的转录组失调。这在最有症状的CD8 + stat3突变患者中更为明显,这是一种与其他疾病亚组分离的独特生物实体。特别是,CD8 + stat3突变的病例表现出广泛的基因下调,最终导致增殖和细胞周期途径的去抑制。在CD8 + stat3突变病例中上调的基因中,我们发现了VCAM1、转录抑制因子EZH2和p53调节因子MDM2原癌基因,以及与白血病发生相关的PVT1上调,这是白血病T-LGLs中首次报道的长链非编码RNA改变。STAT5B突变对T-LGLL转录组的影响更为有限,PIM1丝氨酸/苏氨酸激酶原癌基因的过表达被确定为STAT5B突变的CD4 + T-LGLL最相关的特征之一。这项研究显著推进了我们对T-LGLL发病机制的理解,揭示了该疾病不同分子亚型中新的致癌机制。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Transcriptomic landscape of CD8+ and CD4 + T-LGL leukemia revealed the distinct impact of STAT3 and STAT5B activating mutations
The biological basis of the high clinical heterogeneity of T-LGL Leukemia (T-LGLL) is not completely understood and effective therapies for this disease are lacking. Through RNA-Sequencing of purified T-LGLs we reveal gene expression profiles and pathway dysregulations in the major patient subgroups, defined by CD8+ or CD4+ phenotype and STAT3/STAT5B mutational status. Overall, T-LGLL patients exhibited a marked transcriptome dysregulation compared to controls. This was more pronounced in the most symptomatic CD8 + STAT3-mutated patients, which emerged as a distinct biological entity, separated from the other disease subgroups. Particularly, CD8 + STAT3-mutated cases displayed extensive down-regulation of genes, ultimately resulting in the de-repression of proliferation and cell cycle pathways. Among genes up-regulated in CD8 + STAT3-mutated cases we found VCAM1, the transcriptional repressor EZH2 and the p53-regulator MDM2 proto-oncogene, as well as the leukemogenesis-associated PVT1 up-regulation, representing the first report of a long-non-coding RNA alterations in leukemic T-LGLs. The impact of STAT5B mutations on T-LGLs transcriptome was more limited and the overexpression of the PIM1 serine/threonine kinase proto-oncogene was identified as one of the most relevant features of STAT5B-mutated CD4 + T-LGLL. This study significantly advances our understanding of T-LGLL pathogenesis, uncovering new oncogenic mechanisms within the distinct molecular subtypes of the disease.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Leukemia
医学-血液学
CiteScore
18.10
自引率
3.50%
发文量
270
审稿时长
3-6 weeks
期刊介绍:
Title: Leukemia
Journal Overview:
Publishes high-quality, peer-reviewed research
Covers all aspects of research and treatment of leukemia and allied diseases
Includes studies of normal hemopoiesis due to comparative relevance
Topics of Interest:
Oncogenes
Growth factors
Stem cells
Leukemia genomics
Cell cycle
Signal transduction
Molecular targets for therapy
And more
Content Types:
Original research articles
Reviews
Letters
Correspondence
Comments elaborating on significant advances and covering topical issues
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


