用纵向观测资料估计个体化治疗效果的变分时间解方正网络
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:通过利用现实世界的电子健康记录(EHR)数据,本研究旨在评估纵向观察环境下的个性化治疗效果(ITE),以推进个性化医疗,解决现实世界临床场景中经常观察到的关键挑战,并提出统计挑战,包括隐藏的混杂因素和动态治疗方案。方法我们提出了变分时间反创立者网络(VTDNet),这是一种新的框架,旨在利用基于变分循环变压器的自编码器来解释时变隐藏混淆。具体而言,VTDNet包括三个关键组件:捕获隐藏表示的时序编码器-解码器结构,捕获多种治疗之间相互依赖关系的治疗块,以及预测事实和反事实结果的潜在结果块。我们使用一个合成数据集和两个真实世界的数据集来评估拟议框架的有效性:MIMIC-III,一个专注于重症监护区的EHR数据集,和NACC,强调神经退行性疾病,使用标准化协议从阿尔茨海默病研究中心(ADRC)临床核心的参与者中收集。结果在合成数据集上的实验结果表明,在不同的混杂程度下,合成数据集的准确率都很高。在现实世界的EHR数据集上,与现有的最先进的方法相比,VTDNet在估计异构效应方面实现了更低的均方根误差、平均绝对误差和影响函数精度。结论提出的VTDNet提供了一个稳健的框架来估计纵向环境下的个性化治疗效果,有效地适应不规则时间点和高维数据,同时通过深度生成方法解决隐藏的混杂因素。它在推进个性化医疗和支持真实世界证据生成方面具有巨大潜力。未来的工作将致力于将VTDNet扩展到持续治疗场景,如剂量-反应分析,以进一步扩大其在临床实践中的适用性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
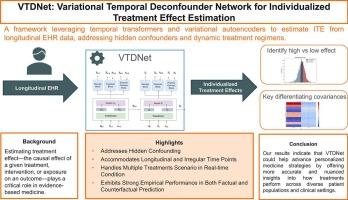
Variational temporal deconfounder network for individualized treatment effect estimation with longitudinal observational data
Objective
By leveraging real-world electronic health record (EHR) data, this study set out to estimate individualized treatment effects (ITE) in longitudinal observational settings to advance personalized medicine, addressing key challenges that are often observed in real-world clinical scenarios and pose statistical challenges, including hidden confounding and dynamic treatment regimens.
Methods
We propose the Variational Temporal Deconfounder Network (VTDNet), a novel framework designed to account for time-varying hidden confounding using a variational recurrent transformer-based autoencoder. Specifically, VTDNet comprises three critical components: a temporal Encoder-Decoder structure to capture hidden representation, a Treatment Block that captures interdependencies among multiple treatments, and a Potential Outcome Block that predicts both factual and counterfactual outcomes. We assess the effectiveness of the proposed framework using a synthetic dataset and two real-world datasets: MIMIC-III, an EHR dataset focusing on intensive care settings, and NACC, emphasizing neurodegenerative disease, collected using a standardized protocol from participants enrolled in Alzheimer’s Disease Research Center (ADRC) clinical cores.
Results
Experimental results on the synthetic dataset demonstrate superior accuracy under varying levels of confounding. On real-world EHR datasets, VTDNet achieves lower root mean squared error, mean absolute error, and influence function precision in the estimation of heterogeneous effects compared to existing state-of-the-art methods.
Conclusion
The proposed VTDNet offers a robust framework for estimating individualized treatment effects in longitudinal settings, effectively accommodating irregular time points and high-dimensional data while addressing hidden confounders through a deep generative approach. It holds significant potential to advance personalized medicine and support real-world evidence generation. Future work will aim to extend VTDNet to continuous treatment scenarios, such as dose–response analysis, to further broaden its applicability in clinical practice.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


