肾上皮样血管平滑肌脂肪瘤和良性血管平滑肌脂肪瘤的基因组图谱和分子进化轨迹
IF 7.7
2区 医学
Q1 Biochemistry, Genetics and Molecular Biology
引用次数: 0
摘要
肾血管平滑肌脂肪瘤(AML)包括良性变种(脂肪瘤型[L-AML],肌瘤型[M-AML])和上皮样AML (eAML),这是一种潜在的恶性亚型,与侵袭行为相关。虽然TSC1/TSC2突变很常见,但eAML发病机制的分子驱动因素尚不清楚。对35份AML样本(15份eAML, 10份L-AML, 10份M-AML)进行了全外显子组测序(WES),并进行了匹配的种系对照。使用OncodriveCLUST和MutSigCV鉴定驱动基因。结合表达谱和生存分析对71例FFPE样本进行验证。这一发现表明,TSC2是AML中最常见的突变致病基因,突变率为69%。TSC2、POLDIP2、NEFH和MUC2是AML亚型的潜在驱动基因,而RHPN2、ASXL1、TOP3B和USP35表现出亚型特异性突变。值得注意的是,在AML变体中观察到明显的细胞遗传学畸变,包括3p26.3, 5p13.1, 6p22.1和11p11.11缺失。克隆进化分析表明,l-AML和eAML,以及M-AML和eAML可能起源于一个共同的祖先克隆,保留了早期突变,并在分化后获得了额外的改变。低TSC2/POLDIP2和高NEFH/MUC2表达与eAML患者的有利生存相关。重要的是,较低的TSC2/POLDIP2表达也预示着依维莫司治疗的更高应答率。总之,我们的研究全面描述了肾性AML亚型之间的基因组差异和进化轨迹,建立了TSC2、POLDIP2、NEFH和MUC2作为预后生物标志物和治疗预测因子,促进了肾性AML治疗的精准医学。本文章由计算机程序翻译,如有差异,请以英文原文为准。
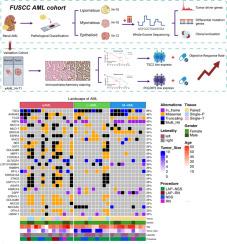
Genomic landscape and molecular evolutionary trajectories of renal epithelioid angiomyolipoma and benign angiomyolipoma
Renal angiomyolipoma (AML) encompasses benign variants (lipomatous [L-AML], myomatous [M-AML]) and epithelioid AML (eAML), a potentially malignant subtype associated with aggressive behavior. While TSC1/TSC2 mutations are frequent, the molecular drivers underlying eAML pathogenesis remain unclear. Whole-exome sequencing (WES) was performed on 35 AML samples (15 eAML, 10 L-AML, 10 M-AML) with matched germline controls. Driver genes were identified using OncodriveCLUST and MutSigCV. Validation was conducted on 71 FFPE samples integrating expression profiling and survival analysis. The finding suggested that TSC2 emerged as the most frequently mutated pathogenic gene in AML, exhibiting a mutation rate of 69 %. TSC2, POLDIP2, NEFH, and MUC2 emerged as potential driver genes across AML subtypes, whereas RHPN2, ASXL1, TOP3B, and USP35 showed subtype-specific mutations. Notably, distinct cytogenetic aberrations were observed among AML variants, including deletions at 3p26.3, 5p13.1, 6p22.1, and 11p11.11. Clonal evolution analysis suggested that l-AML and eAML, as well as M-AML and eAML, may originate from a common ancestral clone, retaining early mutations and acquiring additional alterations post-divergence. Low TSC2/POLDIP2 and high NEFH/MUC2 expression correlated with favorable survival in eAML patients. Importantly, lower TSC2/POLDIP2 expression also predicted superior response rates to Everolimus therapy. In conclusion, our study comprehensively delineates genomic distinctions and evolutionary trajectories among renal AML subtypes, establishing TSC2, POLDIP2, NEFH, and MUC2 as prognostic biomarkers and therapeutic predictors, facilitating precision medicine in eAML management.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Neoplasia
医学-肿瘤学
CiteScore
9.20
自引率
2.10%
发文量
82
审稿时长
26 days
期刊介绍:
Neoplasia publishes the results of novel investigations in all areas of oncology research. The title Neoplasia was chosen to convey the journal’s breadth, which encompasses the traditional disciplines of cancer research as well as emerging fields and interdisciplinary investigations. Neoplasia is interested in studies describing new molecular and genetic findings relating to the neoplastic phenotype and in laboratory and clinical studies demonstrating creative applications of advances in the basic sciences to risk assessment, prognostic indications, detection, diagnosis, and treatment. In addition to regular Research Reports, Neoplasia also publishes Reviews and Meeting Reports. Neoplasia is committed to ensuring a thorough, fair, and rapid review and publication schedule to further its mission of serving both the scientific and clinical communities by disseminating important data and ideas in cancer research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


