基于超声图像的结直肠肿瘤三维分割的深度学习策略
IF 4.2
3区 计算机科学
Q2 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
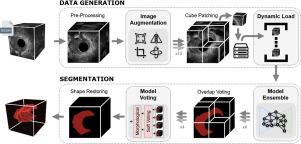
结直肠癌仍然是世界范围内癌症相关死亡的主要原因,这突出了对准确和高效诊断工具的需求。虽然深度学习在医学成像方面显示出前景,但其在经直肠超声结肠直肠肿瘤分割中的应用仍未得到充分探索。目前,病变分割是依靠临床医生的专业知识手动进行的,导致各个治疗中心的差异很大。为了克服这一限制,我们提出了一种新的策略,解决了实际挑战和技术限制,特别是在数据可用性有限的情况下,为从超声成像中准确分割结肠直肠肿瘤提供了一个强大的框架。我们评估了八种最先进的模型,包括卷积神经网络和基于变压器的架构,并引入了适合领域的预处理和后处理技术,如数据增强、修补和集成,以提高分割性能,同时降低计算成本。与基线模型相比,DICE评分平均提高0.423绝对分(+107%),我们的研究结果证明了我们的建议在提高基于超声的结直肠癌诊断的准确性和可靠性方面的潜力,为临床可行的ai驱动解决方案铺平了道路。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A deep learning strategy for the 3D segmentation of colorectal tumors from ultrasound imaging
Colorectal cancer remains a leading cause of cancer-related mortality worldwide, highlighting the need for accurate and efficient diagnostic tools. While Deep Learning has shown promise in medical imaging, its application to transrectal ultrasound for colorectal tumor segmentation remains underexplored. Currently, lesion segmentation is performed manually, relying on clinician expertise and leading to significant variability across treatment centers. To overcome this limitations, we propose a novel strategy that addresses both practical challenges and technical constraints, particularly in scenarios with limited data availability, offering a robust framework for accurate 3D colorectal tumor segmentation from ultrasound imaging. We evaluate eight state-of-the-art models, including convolutional neural networks and transformer-based architectures, and introduce domain-tailored pre- and post-processing techniques such as data augmentation, patching and ensembling to enhance segmentation performance while reducing computational cost. Leading to an average improvement in term of DICE score of 0.423 absolute points (+107%), compared to baseline models, our findings demonstrate the potential of our proposal to improve the accuracy and reliability of ultrasound-based diagnostics for colorectal cancer, paving the way for clinically viable AI-driven solutions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Image and Vision Computing
工程技术-工程:电子与电气
CiteScore
8.50
自引率
8.50%
发文量
143
审稿时长
7.8 months
期刊介绍:
Image and Vision Computing has as a primary aim the provision of an effective medium of interchange for the results of high quality theoretical and applied research fundamental to all aspects of image interpretation and computer vision. The journal publishes work that proposes new image interpretation and computer vision methodology or addresses the application of such methods to real world scenes. It seeks to strengthen a deeper understanding in the discipline by encouraging the quantitative comparison and performance evaluation of the proposed methodology. The coverage includes: image interpretation, scene modelling, object recognition and tracking, shape analysis, monitoring and surveillance, active vision and robotic systems, SLAM, biologically-inspired computer vision, motion analysis, stereo vision, document image understanding, character and handwritten text recognition, face and gesture recognition, biometrics, vision-based human-computer interaction, human activity and behavior understanding, data fusion from multiple sensor inputs, image databases.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


