医疗保健数据库研究中可扩展特征工程和超高维混杂调整的自然语言处理。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
背景:为了改善医疗数据库研究中的混杂控制,数据驱动算法可以通过经验识别和调整大量暴露前变量,这些变量间接捕获了未测量混杂因素(“代理”混杂因素)的信息。目前用于高维代理调整的方法不能利用电子健康记录(EHRs)中的自由文本注释。无监督自然语言处理(NLP)技术可以扩展到从非结构化票据中生成大量结构化特征。目的:评估用大量NLP生成的特征补充理赔数据分析对高维代理调整的影响。方法:我们将医疗保险索赔与电子病历数据联系起来,生成三个队列,比较不同类别的药物对6个月心血管结局风险的影响。我们使用各种NLP方法从自由文本EHR笔记中生成结构化特征,并使用最小绝对收缩和选择算子(LASSO)回归来拟合几种倾向评分(PS)模型,这些模型包括不同的协变量集作为候选预测因子。协变量集包括仅由索赔数据生成的特征,以及索赔数据加上nlp生成的EHR特征。结果:将索赔代码和nlp生成的EHR特征作为候选预测因子,改善了总体协变量平衡,标准化差异为 结论:用大量nlp生成的特征补充行政索赔,用于超高维代理混杂因素调整,改善了总体协变量平衡,并可能在捕获混杂因素信息方面提供适度的好处。本文章由计算机程序翻译,如有差异,请以英文原文为准。
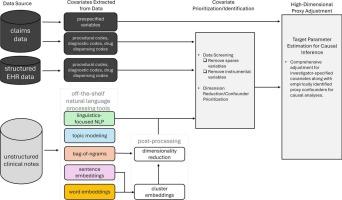
Natural language processing for scalable feature engineering and ultra-high-dimensional confounding adjustment in healthcare database studies
Background
To improve confounding control in healthcare database studies, data-driven algorithms may empirically identify and adjust for large numbers of pre-exposure variables that indirectly capture information on unmeasured confounding factors (‘proxy’ confounders). Current approaches for high-dimensional proxy adjustment do not leverage free-text notes from electronic health records (EHRs). Unsupervised natural language processing (NLP) technology can scale to generate large numbers of structured features from unstructured notes.
Objective
To assess the impact of supplementing claims data analyses with large numbers of NLP generated features for high-dimensional proxy adjustment.
Methods
We linked Medicare claims with EHR data to generate three cohorts comparing different classes of medications on the 6-month risk of cardiovascular outcomes. We used various NLP methods to generate structured features from free-text EHR notes and used least absolute shrinkage and selection operator (LASSO) regression to fit several propensity score (PS) models that included different covariate sets as candidate predictors. Covariate sets included features generated from claims data only, and claims data plus NLP-generated EHR features.
Results
Including both claims codes and NLP-generated EHR features as candidate predictors improved overall covariate balance with standardized differences being < 0.1 for all variables. While overall balance improved, the impact on estimated treatment effects was more nuanced with adjustment for NLP-generated features moving effect estimates further in the expected direction in two of the empirical studies but had no impact on the third study.
Conclusion
Supplementing administrative claims with large numbers of NLP-generated features for ultra-high-dimensional proxy confounder adjustment improved overall covariate balance and may provide a modest benefit in terms of capturing confounder information.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


