揭示蜂胶对单核增生李斯特菌转录调控因子prfA的潜在靶向活性,以对抗李斯特菌病
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
单核细胞增生李斯特菌是世界范围内爆发李斯特菌病的病原体,由于耐药性和死亡率的出现,李斯特菌引起了人们的高度警惕。一种转录调节因子(prfA)调节单核增生乳杆菌的所有毒力级联反应,使其成为许多药物分子的独特和假定的靶点。蜂胶是自然界隐藏的宝藏,是一种复杂的异质混合物,含有许多植物源次生代谢物。蜜蜂可能会产生一些活性化合物,利用它们的生物活性潜力是治疗各种疾病的新兴兴趣。在此背景下,本研究的重点是评价蜂胶的主要生物活性化合物对单核增生乳杆菌的推定靶标(prfA)的靶向作用。在各种蜂胶化合物中,选择了75种配体与prfA的A链对接。首先,在薛定谔套件2023_3中使用QikProp v_5.8评估配体的药代动力学性质。所得蜂胶化合物的对接分数在−13.022 ~−5.171 kcal/mol范围内,所有药动学参数均符合要求。对配体70(−13.022 kcal/mol)和配体39(−12.58 kcal/mol)等负对接分数较高的化合物进行了分子动力学模拟研究,利用Maestro软件v_11.8中嵌入的Desmond v_5.6包确定了它们在100 ns模拟过程中的结合稳定性,然后用prime MM/GBSA计算了配体-受体配合物的结合自由能。所有参数都设想了prfA(靶蛋白)活性位点配体分子的稳定性,以抑制单核细胞增生乳杆菌在宿主中的致病性。综上所述,蜂胶化合物通过积极抑制转录调控因子(prfA)的活性,协同作用于单核增生乳杆菌,以对抗李斯特菌病的爆发,从而确保食品安全和公众健康。本文章由计算机程序翻译,如有差异,请以英文原文为准。
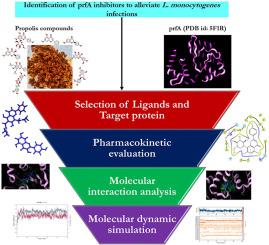
Unraveling the potential targeting activity of propolis against transcriptional regulator prfA from Listeria monocytogenes to combat listeriosis
Listeria monocytogenes is a causative agent for pernicious listeriosis outbreaks worldwide and it has attained more alertness due to the emergence of resistance and mortality rate. A transcriptional regulator (prfA) regulates all the virulence cascades in L. monocytogenes making it a unique and putative target for many drug molecules. Propolis, nature's hidden treasure, is a complex and heterogeneous mixture that contains many secondary metabolites of plant origin. Bees might produce some active compounds and harnessing their bioactive potential is a burgeoning interest to treat various illnesses. In this context, the present study focuses on evaluating the targeted action of the major bioactive compounds of propolis against the putative target (prfA) of L. monocytogenes. Among various propolis compounds, 75 ligands were selected and docked with the A chain of prfA. Initially, the pharmacokinetic properties of the ligands were evaluated using QikProp v_5.8 in the Schrodinger suite 2023_3. All the pharmacokinetic parameters were satisfied with the selected propolis compounds and the docking score of the compounds obtained was in the range of −13.022 to −5.171 kcal/mol. The compounds with high negative docking scores, such as Ligand 70 (−13.022 kcal/mol) and Ligand 39 (−12.58 kcal/mol) were subjected to molecular dynamics simulation studies to determine their binding stability for a 100 ns simulation course using Desmond v_5.6 packages embedded in the Maestro software v_11.8, followed by the binding free energies of the ligand-receptor complexes were computed using prime MM/GBSA. All the parameters have envisaged the stability of the ligand molecules at the active site of prfA (target protein) to inhibit L. monocytogenes pathogenicity in the host. In sum, the compounds of propolis synergistically act against L. monocytogenes by actively inhibiting the activity of transcriptional regulators (prfA) to combat listeriosis outbreaks, thus ensuring food safety and public health.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


