ETV6::RUNX1白血病治疗反应的基因组决定因素
IF 13.4
1区 医学
Q1 HEMATOLOGY
引用次数: 0
摘要
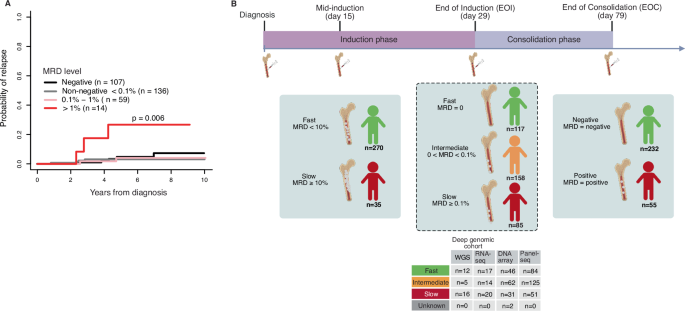
RUNX1白血病是儿童B细胞急性淋巴细胞白血病(B- all)的第二常见亚型。虽然它的复发风险一般较低,但由于其发病率相对较高,B-ALL复发在该亚型中占很大比例。诱导治疗结束时可测量的残留疾病是一个公认的预测治疗结果的生物标志物,而没有基因组生物标志物在临床中常规应用。在这项研究中,我们使用来自ETV6::RUNX1白血病的多组学数据来确定疾病出现时预测治疗反应的基因组特征。在深入表征的亚队列中,我们发现快速反应的病例经常表现出APOBEC突变特征和高细胞周期活性的基因表达特征。相比之下,IGK基因重排在反应缓慢的患者中更为频繁。此外,在转录调节因子和肿瘤抑制基因(INTS1, NF1, TP53)中发现了应答相关突变。拷贝数分析显示,快速应答者更频繁地缺失chr12 p臂,导致影响诱导治疗应答相关基因的转录组变化(KRAS, FKBP4),而较短的chr12增加在缓慢应答者中更为常见。已确定的治疗敏感性的遗传和转录组标记为改善疾病的表现分类铺平了道路,潜在地改善了临床结果。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Genomic determinants of therapy response in ETV6::RUNX1 leukemia
ETV6::RUNX1 leukemia is the second most common subtype of childhood B cell acute lymphoblastic leukemia (B-ALL). Although it generally has a low relapse risk, a significant proportion of B-ALL relapses occur within this subtype due to its relatively high incidence. Measurable residual disease at the end of induction therapy is a well-established biomarker predicting treatment outcomes, while no genomic biomarkers are routinely applied in clinics. In this study, we used multiomic data from ETV6::RUNX1 leukemias to identify genomic features predictive of therapy response at disease presentation. In the deeply characterized sub-cohort we discovered that fast-responding cases frequently exhibited the APOBEC mutational signature and the gene expression signature of high cell cycle activity. In contrast, rearrangements of IGK genes were more frequent in slow responders. Additionally, response-related mutations were identified in transcriptional regulators and tumor suppressor genes (INTS1, NF1, TP53). Copy number analysis revealed that fast responders harbored more frequent deletions of chr12 p-arm, leading to transcriptomic changes affecting genes associated with induction therapy response (KRAS, FKBP4), while a shorter gain in chr12 was more common in slow responders. The identified genetic and transcriptomic markers of treatment sensitivity pave the way for improved disease classification at presentation, potentially improving clinical outcomes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Leukemia
医学-血液学
CiteScore
18.10
自引率
3.50%
发文量
270
审稿时长
3-6 weeks
期刊介绍:
Title: Leukemia
Journal Overview:
Publishes high-quality, peer-reviewed research
Covers all aspects of research and treatment of leukemia and allied diseases
Includes studies of normal hemopoiesis due to comparative relevance
Topics of Interest:
Oncogenes
Growth factors
Stem cells
Leukemia genomics
Cell cycle
Signal transduction
Molecular targets for therapy
And more
Content Types:
Original research articles
Reviews
Letters
Correspondence
Comments elaborating on significant advances and covering topical issues
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


