基于秩的稀疏神经网络混合结果联合建模。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:在过去的几十年里,高通量谱分析得到了广泛的应用,在癌症研究、生存分析和其他生物医学研究中取得了重大进展。虽然已经开发了许多方法来识别重要特征并构建预测模型,但由于高维数和小样本量导致信息不足,生物医学研究经常面临挑战,这往往导致识别和预测精度不理想。方法:在本文中,我们提出了一个基于秩的稀疏神经网络,有效地利用混合结果的信息,特别是纳入生存数据。该方法考虑了结果与高维协变量之间的未知关系,而许多传统方法是建立在参数框架上的。提出了一种新的损失函数来解决梯度不平衡问题并适应混合结果。开发了一个稀疏层来实现惩罚方法,使重要变量的识别成为可能。结果:我们进行了大量的仿真研究,表明所提出的方法是有效的,具有广泛的适用性。对皮肤黑色素瘤(SKCM)的分析证明了我们提出的方法的竞争性能。结论:该方法有效地对混合结果(包括生存数据)进行建模,并选择重要特征,有利于癌症和基因组研究等生物医学研究。本文章由计算机程序翻译,如有差异,请以英文原文为准。
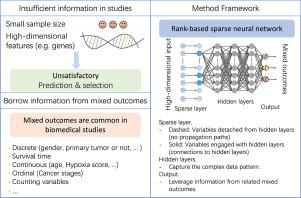
Joint modeling of mixed outcomes using a rank-based sparse neural network
Objective:
In the past few decades, high-throughput profiling has been extensively conducted, leading to significant advancements in cancer research, survival analysis, and other biomedical studies. While many methods have been developed to identify important features and construct predictive models, biomedical research often faces challenges due to insufficient information caused by high dimensionality and small sample sizes, which frequently lead to unsatisfactory identification and prediction accuracy.
Methods:
In this paper, we propose a rank-based sparse neural network that efficiently leverages information from mixed outcomes, particularly incorporating survival data. The proposed method accounts for unknown relationships between outcomes and high-dimensional covariates, whereas many traditional methods are built on a parametric framework. A novel loss function is derived to address the gradient imbalance issue and accommodate mixed outcomes. A sparse layer is developed to implement the penalization method, enabling the identification of important variables.
Results:
We conducted extensive simulation studies, showing that the proposed method is effective and broadly applicable. The analysis of skin cutaneous melanoma (SKCM) demonstrates the competitive performance of our proposed method.
Conclusion:
The proposed method effectively models mixed outcomes (including survival data) and selects important features, which is beneficial for biomedical studies like cancer and genomic research.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


