针对ESKAPE病原体的广谱疫苗的计算机开发
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
耐药ESKAPE病原体(粪肠球菌、金黄色葡萄球菌、肺炎克雷伯菌、鲍曼不动杆菌、铜绿假单胞菌和阴沟肠杆菌)严重限制了重症感染的治疗选择,从而增加了传染病的严重程度和死亡率,这是一个重大的全球卫生挑战。疫苗接种作为一种预防措施对于大幅度减少细菌感染至关重要,并且对耐抗生素细菌可能有效。本研究展示了一种基于表位的疫苗的设计,该疫苗能够中和ESKAPE病原体中存在的共享抗原决定因子。对ESKAPE病原菌进行泛基因组分析,提取核心蛋白质组。这种方法有助于反向疫苗学分析,以确定该细菌群中的抗原蛋白。该研究发现,在孔蛋白OmpA、OprD和TolC以及胶原结合粘接剂Acm和Cna中存在类似的结构。然后利用这些蛋白预测t细胞和b细胞表位,选择那些具有最佳物理化学性质、抗原性、非过敏原性和缺乏毒性的蛋白。此外,位于抗原表面且能够与HLA分子偶联的表位被优先考虑。在这种计算方法中,我们设计了一个包含佐剂RS09、TLR4激动剂和通过连接物连接的免疫原性表位的结构。我们通过分子对接和分子动力学模拟来评估它们与免疫系统模式识别受体相互作用的稳定性。计算机免疫模拟表明,该疫苗可触发体液和细胞介导的免疫反应。由此产生的结构可能代表了一种有效和安全的候选疫苗,以预防ESKAPE组引起的感染。本文章由计算机程序翻译,如有差异,请以英文原文为准。
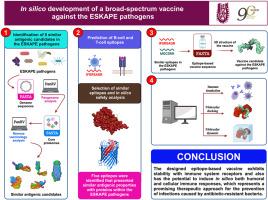
In silico development of a broad-spectrum vaccine against ESKAPE pathogens
Antimicrobial-resistant ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter cloacae) have significantly restricted therapeutic alternatives for critical infections, consequently contributing to increase the severity and mortality of infectious illnesses that represent a significant global health challenge. Vaccination as a preventive measure can be crucial in substantially reducing bacterial infections and is potentially effective against antibiotic-resistant bacteria. This study shows the design of an epitope-based vaccine capable of neutralizing shared antigenic determinants present among the ESKAPE pathogens. The pangenome of the ESKAPE pathogens was analyzed to extract the core proteome. This approach facilitated reverse vaccinology analysis to identify antigenic proteins within this bacterial group. The study revealed similar structures in porins OmpA, OprD, and TolC, as well as the collagen-binding adhesins Acm and Cna. These proteins were then utilized to predict T-cell and B-cell epitopes, selecting those with their best physicochemical properties, antigenicity, non-allergenicity, and lack of toxicity. Additionally, epitopes located on the surface of the antigens and capable of coupling with HLA molecules were prioritized. In this computational approach, we engineered a construct incorporating the adjuvant RS09, a TLR4 agonist, and immunogenic epitopes connected by linkers. We assessed the stability of their interaction with pattern recognition receptors of the immune system through molecular docking and molecular dynamics simulations. The in silico immune simulation demonstrated that the vaccine could trigger humoral and cell-mediated immune responses. The resulting construct potentially represents an effective and safe vaccine candidate to prevent infections caused by the ESKAPE group.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


