AnalyzAIRR:一个用户友好的指导工作流,用于AIRR数据分析
引用次数: 0
摘要
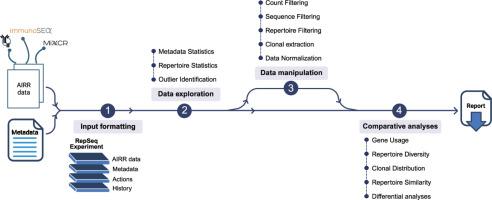
对大量适应性免疫受体(AIRR)的分析使我们能够理解正常和病理条件下的免疫反应。然而,AIRR的复杂性需要先进的、专门的方法来提取有意义的生物学见解。这些复杂的方法往往给生物信息学专业知识有限的研究人员带来挑战,阻碍了获得全面的免疫系统分析。为了应对这一挑战,我们开发了AnalyzAIRR,这是一个符合AIRR标准的R包,可以实现先进的批量AIRR测序数据。该工具集成了适用于不同粒度级别的最先进的统计和可视化方法。它为一般数据探索、过滤和操作提供了一个平台,并对AIRR数据集进行了深入的交叉比较,旨在回答特定的生物学问题。我们使用已发表的来自三个不同T细胞亚群的18个T细胞受体的小鼠数据集来说明AnalyzAIRR的功能。在进行对比分析之前,我们首先在一组样品中检测并去除了一种主要污染物。随后的跨样本分析揭示了与各自细胞表型一致的库多样性差异,以及研究亚群之间的库收敛性差异。AnalyzAIRR的分析指标集集成到Shiny的web应用程序中,并辅以教程来帮助用户进行分析策略,使其对生物信息学背景很少或没有背景的生物学家来说都是用户友好的。本文章由计算机程序翻译,如有差异,请以英文原文为准。
AnalyzAIRR: A user-friendly guided workflow for AIRR data analysis
The analysis of bulk adaptive immune receptor repertoires (AIRR) enables the understanding of immune responses in both normal and pathological conditions. However, the complexity of AIRR calls for advanced, specialized methods to extract meaningful biological insights. These sophisticated approaches often present challenges for researchers with limited bioinformatics expertise, hindering access to comprehensive immune system analysis. To address this challenge, we developed AnalyzAIRR, an AIRR-compliant R package enabling advanced bulk AIRR sequencing data. The tool integrates state-of-the-art statistical and visualization methods applicable at various levels of granularity. It offers a platform for general data exploration, filtering and manipulation, and in-depth cross-comparisons of AIRR datasets, aimed at answering specific biological questions. We illustrate AnalyzAIRR functionalities using a published murine dataset of 18 T-cell receptor repertoires from three diferrent T cell subsets. We first detected and removed a major contaminant in a group of samples, before proceeding with the comparative analysis. Subsequent cross-sample analysis revealed differences in repertoire diversity that aligned with the respective cell phenotypes, and in repertoire convergence among the studied subsets. AnalyzAIRR’s set of analytical metrics is integrated into a Shiny web application and complemented with a tutorial to help users in their analytical strategy, making it user-friendly for biologists with little or no background in bioinformatics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Immunoinformatics (Amsterdam, Netherlands)
Immunology, Computer Science Applications
自引率
0.00%
发文量
0
审稿时长
60 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


