用Cytoland对地标细胞器进行鲁棒虚拟染色
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
具有里程碑意义的细胞器(如细胞核、核仁、细胞膜、核膜和脂滴)的相关活细胞成像对系统细胞生物学和药物发现至关重要。然而,仅用分子标记实现这一目标仍然具有挑战性。利用深度神经网络从无标记图像中对多个细胞器和细胞状态进行虚拟染色是一种新兴的解决方案。虚拟染色为成像分子传感器、图像操作或其他任务释放了光谱。目前用于标记细胞器虚拟染色的方法通常在成像、培养条件和细胞类型存在令人讨厌的变化时失败。在这里,我们用Cytoland来解决这个问题,Cytoland是一系列模型,用于跨不同成像参数、细胞状态和类型的里程碑细胞器的鲁棒虚拟染色。这些模型使用灵活的卷积架构(UNeXt2)和受光学显微镜图像形成启发的增强进行自监督和监督预训练。细胞体模型能够在一系列成像条件下对多种细胞类型的细胞核和膜进行虚拟染色,包括人类细胞系、斑马鱼神经基质、诱导多能干细胞(iPSCs)和ipsc衍生的神经元。我们使用从虚拟和实验染色的细胞核和膜获得的强度,分割和应用特定测量来评估模型。这些模型挽救了缺失的标签,纠正了不均匀的标签,减轻了光漂白。我们共享多个预训练模型、用于训练、推理和部署的开源软件(VisCy)以及数据集。本文章由计算机程序翻译,如有差异,请以英文原文为准。
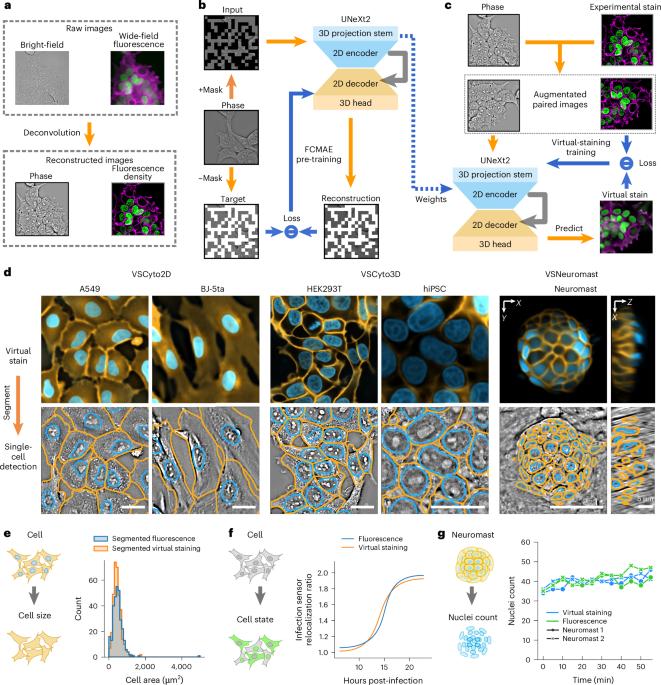
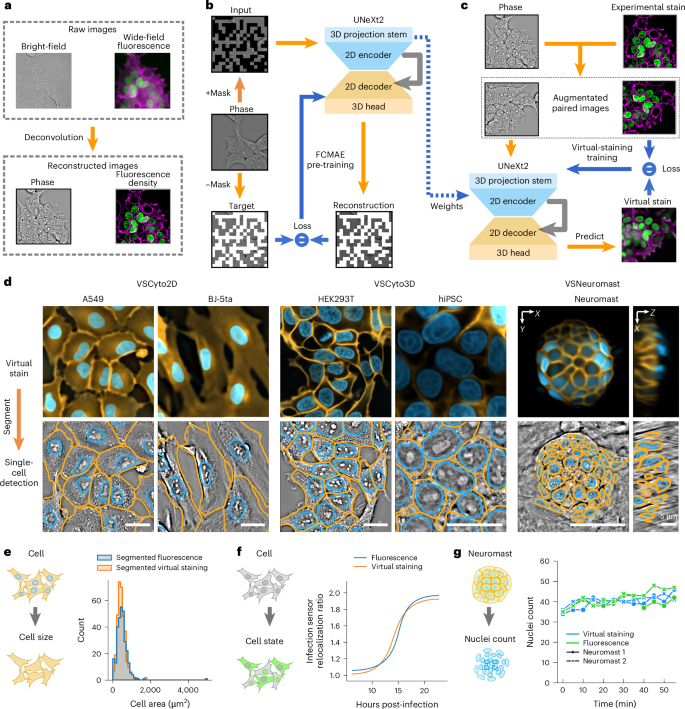
Robust virtual staining of landmark organelles with Cytoland
Correlative live-cell imaging of landmark organelles—such as nuclei, nucleoli, cell membranes, nuclear envelope and lipid droplets—is critical for systems cell biology and drug discovery. However, achieving this with molecular labels alone remains challenging. Virtual staining of multiple organelles and cell states from label-free images with deep neural networks is an emerging solution. Virtual staining frees the light spectrum for imaging molecular sensors, photomanipulation or other tasks. Current methods for virtual staining of landmark organelles often fail in the presence of nuisance variations in imaging, culture conditions and cell types. Here we address this with Cytoland, a collection of models for robust virtual staining of landmark organelles across diverse imaging parameters, cell states and types. These models were trained with self-supervised and supervised pre-training using a flexible convolutional architecture (UNeXt2) and augmentations inspired by image formation of light microscopes. Cytoland models enable virtual staining of nuclei and membranes across multiple cell types—including human cell lines, zebrafish neuromasts, induced pluripotent stem cells (iPSCs) and iPSC-derived neurons—under a range of imaging conditions. We assess models using intensity, segmentation and application-specific measurements obtained from virtually and experimentally stained nuclei and membranes. These models rescue missing labels, correct non-uniform labelling and mitigate photobleaching. We share multiple pre-trained models, open-source software (VisCy) for training, inference and deployment, and the datasets. Ziwen Liu et al. report Cytoland, an approach to train robust models to virtually stain landmark organelles of cells and address the generalization gap of current models. The training pipeline, models and datasets are shared under open-source permissive licences.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


