预测药物-药物相互作用:基于gcn协同过滤的深度学习方法
IF 6.2
2区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
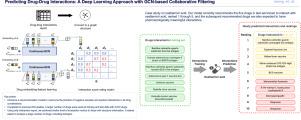
由于与单一疗法相比,联合药物在患者中的使用正在增加。然而,医疗保健提供者在使用联合用药时应继续关注由药物-药物相互作用(ddi)引起的与患者安全相关的潜在风险。虽然直接的物理化学相互作用有助于某些ddi病例,但大多数ddi发生是因为一种药物调节酶,如细胞色素P450,负责代谢另一种药物。因此,与同一家族药物相互作用的药物更有可能通过介导特定酶相互作用。根据推荐具有相似兴趣的用户的技术,我们引入了一种具有图卷积网络(GCN)和协同过滤的人工智能推荐模型,该模型分析相互作用药物的连通性,而不是它们的化学结构。这种方法偏离了典型的分类模型,不需要将未定义的相互作用作为负样本进行采样,允许预测所有未知药物对的潜在相互作用,避免了与选择负相互作用和数据不平衡相关的挑战。我们的方法使用了DrugBank数据库(2022年1月3日发布的5.1.9版本),包含4072种药物和1391790对相互作用的药物。此外,通过5倍验证和使用TWOSIDES数据的外部数据验证,验证了模型的稳健性。值得注意的是,我们的模型的功效完全是通过利用DDI报告来建立的,它提供了一个能够准确预测不同药物类型之间相互作用的通用框架。这个项目的源代码发布在GitHub上(https://github.com/yeonuk-Jeong/DDI-OCF)。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Predicting drug-drug interactions: A deep learning approach with GCN-based collaborative filtering
The use of combination drugs among patients is increasing due to effectiveness compared to monotherapies. However, healthcare providers should continue to be concerned about the potential risks associated with patient safety arising from drug-drug interactions (DDIs) when they use combination drugs. Whereas direct physicochemical interactions contribute to certain cases of DDIs, the majority of DDIs occur because one drug modulates enzymes, such as cytochrome P450, responsible for metabolizing another drug. Therefore, drugs that interact with the same family drugs are more likely to interact with each other by mediating specific enzymes. Adapted from techniques used to recommend users with similar interests, we introduce an AI recommendation model with graph convolutional network (GCN) and collaborative filtering that analyzes the connectivity of interacting drugs rather than their chemical structures. This approach deviates from typical classification models by not requiring sampling of undefined interactions as negative samples, allowing the prediction of potential interactions for all unknown drug pairs, circumventing the challenges associated with selecting negative interactions and data imbalance. Our methodology used the DrugBank database (version 5.1.9 released on January 3, 2022), encompassing 4,072 drugs and 1,391,790 drug pairs with interactions. Furthermore, the robustness of the model was verified through a 5-fold validation and external data validation using TWOSIDES data. Notably, our model’s efficacy is established solely through the exploitation of DDI reports, offering a versatile framework capable of accurately predicting interactions among diverse drug types. The source code for this project is distributed on GitHub (https://github.com/yeonuk-Jeong/DDI-OCF).
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Artificial Intelligence in Medicine
工程技术-工程:生物医学
CiteScore
15.00
自引率
2.70%
发文量
143
审稿时长
6.3 months
期刊介绍:
Artificial Intelligence in Medicine publishes original articles from a wide variety of interdisciplinary perspectives concerning the theory and practice of artificial intelligence (AI) in medicine, medically-oriented human biology, and health care.
Artificial intelligence in medicine may be characterized as the scientific discipline pertaining to research studies, projects, and applications that aim at supporting decision-based medical tasks through knowledge- and/or data-intensive computer-based solutions that ultimately support and improve the performance of a human care provider.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


