利用Eye2Gene多模态成像研究遗传性视网膜疾病的下一代表型
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
遗传性视网膜疾病(IRDs)等罕见眼病的遗传诊断具有挑战性。IRDs是典型的单基因疾病,是全世界儿童和工作年龄成人失明的主要原因。越来越多的人现在成为临床试验的目标,批准的治疗方法也越来越多。然而,获取需要足够早地建立遗传诊断。关键的是,及时确定遗传原因仍然具有挑战性。我们证明了一种深度学习算法Eye2Gene,在ird个体(n = 2,451)的大型多模态成像数据集上进行了训练,并在五个不同临床中心提供的数据上进行了外部验证,为63种最常见的遗传原因的遗传诊断提供了优于专家水平的前五名准确率83.9%。我们证明Eye2Gene的下一代表型可以通过改进ird筛选、表型驱动的变异优先排序和表型空间中的自动相似性匹配来识别新基因,从而提高诊断率。Eye2Gene可以在线访问(app.eye2gene.com)用于研究目的。本文章由计算机程序翻译,如有差异,请以英文原文为准。
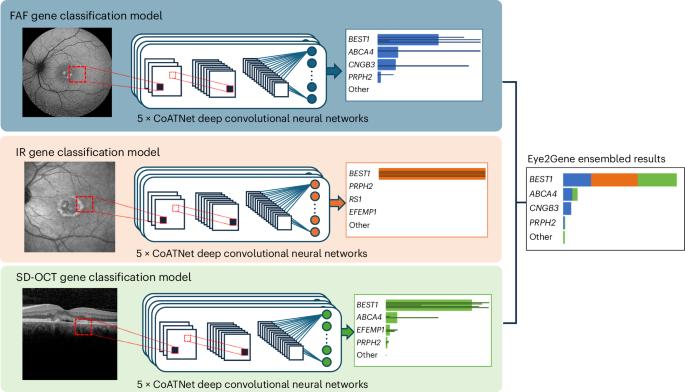

Next-generation phenotyping of inherited retinal diseases from multimodal imaging with Eye2Gene
Rare eye diseases such as inherited retinal diseases (IRDs) are challenging to diagnose genetically. IRDs are typically monogenic disorders and represent a leading cause of blindness in children and working-age adults worldwide. A growing number are now being targeted in clinical trials, with approved treatments increasingly available. However, access requires a genetic diagnosis to be established sufficiently early. Critically, the timely identification of a genetic cause remains challenging. We demonstrate that a deep learning algorithm, Eye2Gene, trained on a large multimodal imaging dataset of individuals with IRDs (n = 2,451) and externally validated on data provided by five different clinical centres, provides better-than-expert-level top-five accuracy of 83.9% for supporting genetic diagnosis for the 63 most common genetic causes. We demonstrate that Eye2Gene’s next-generation phenotyping can increase diagnostic yield by improving screening for IRDs, phenotype-driven variant prioritization and automatic similarity matching in phenotypic space to identify new genes. Eye2Gene is accessible online ( app.eye2gene.com ) for research purposes. Eye2Gene’s next-generation phenotyping of multimodal images increases diagnostic yield for inherited retinal diseases by improving screening, phenotype-driven variant prioritization and automatic similarity matching in phenotypic space to drive gene discovery.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


