统一核酸与蛋白质语言的广义生物学基础模型
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
编码在DNA、RNA和蛋白质中的生物学语言构成了生命的基础,但由于其复杂性,解码仍然具有挑战性。传统的计算方法往往难以整合这些分子之间的信息,从而限制了对生物系统的全面理解。自然语言处理与预训练模型的进展为解释生物语言提供了可能性。在这里,我们介绍LucaOne,一个预先训练的基础模型,训练了来自169,861个物种的核酸和蛋白质序列。通过大规模的数据整合和半监督学习,LucaOne展示了对关键生物学原理的理解,例如dna -蛋白质翻译。通过少量的学习,它有效地理解了分子生物学的核心法则,并在涉及DNA、RNA或蛋白质输入的任务上表现得很有竞争力。我们的研究结果强调了统一基础模型解决复杂生物学问题的潜力,为生物信息学研究提供了一个适应性框架,并增强了对生命复杂性的解释。本文章由计算机程序翻译,如有差异,请以英文原文为准。
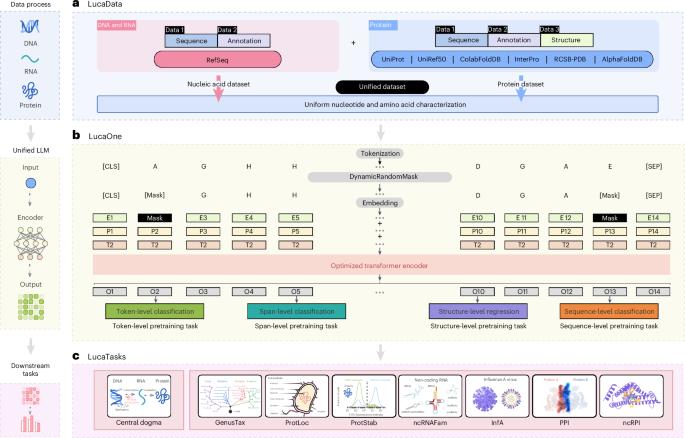

Generalized biological foundation model with unified nucleic acid and protein language
The language of biology, encoded in DNA, RNA and proteins, forms the foundation of life but remains challenging to decode owing to its complexity. Traditional computational methods often struggle to integrate information across these molecules, limiting a comprehensive understanding of biological systems. Advances in natural language processing with pre-trained models offer possibilities for interpreting biological language. Here we introduce LucaOne, a pre-trained foundation model trained on nucleic acid and protein sequences from 169,861 species. Through large-scale data integration and semi-supervised learning, LucaOne shows an understanding of key biological principles, such as DNA–protein translation. Using few-shot learning, it effectively comprehends the central dogma of molecular biology and performs competitively on tasks involving DNA, RNA or protein inputs. Our results highlight the potential of unified foundation models to address complex biological questions, providing an adaptable framework for bioinformatics research and enhancing the interpretation of life’s complexity. He and colleagues develop LucaOne, a biological foundation model pre-trained on nucleic acid and protein sequences from 169,861 species. It shows an emerging understanding of molecular biology’s central dogma, enhancing bioinformatics analysis and helping explore unknown aspects of molecular biology.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


