先进的多光谱光声层析成像(MSOT):用于实时光谱分解的相量分析
IF 6.3
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
背景多光谱光声断层扫描(MSOT)将光学和超声成像相结合,生成高分辨率、分子特异性的图像。通过捕获多个波长的超声波发射,MSOT可以实现组织切片或体积的实时可视化。该技术的特异性依赖于发色团在不同波长发出不同的信号,需要精确的光谱分离。然而,现有的解混方法计算量大,往往难以评估,限制了它们在实时场景中的适用性。方法提出了一种基于相量的快速、直观的MSOT光谱成分定量方法。通过将多光谱数据投影到二维相量平面上,该方法可以直接可视化光谱成分并快速检测意外信号或伪影。该方法通过带有流动血液的光学表征实验幻影进行了验证,并与线性混合模型(LMM)进行了比较。然后将其应用于克罗恩病患者和健康对照者的临床MSOT图像。结果相量法有助于识别伪影和光谱异常,显著提高了MSOT数据的可解释性。在处理速度方面,它也优于传统的解混算法,使其适合于实时应用。在临床数据集中,该方法揭示了克罗恩病患者和健康个体之间不同的光谱模式,突出了其检测生物复杂系统变化和差异的能力。结论相量法作为MSOT光谱解混的有力工具,在数据解释上既快速又清晰。它的实时性和诊断潜力支持MSOT在非侵入性疾病表征中的广泛临床应用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
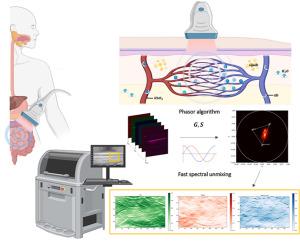
Advancing multispectral optoacoustic tomography (MSOT): Phasor analysis for real-time spectral unmixing
Background
Multispectral optoacoustic tomography (MSOT) merges optical and ultrasound imaging to generate high-resolution, molecularly specific images. By capturing ultrasound emissions at multiple wavelengths, MSOT enables real-time visualization of tissue slices or volumes. The technique's specificity relies on chromophores emitting distinct signals across different wavelengths, requiring accurate spectral unmixing. However, existing unmixing methods are computationally demanding and often difficult to evaluate, limiting their applicability in real-time scenarios.
Method
We present a phasor-based approach as a fast and intuitive solution for spectral component quantification in MSOT. By projecting multispectral data onto a two-dimensional phasor plane, this method enables direct visualization of spectral components and rapid detection of unexpected signals or artifacts. The approach was validated using optically characterized experimental phantoms with flowing blood, allowing comparison with linear mixing models (LMM). It was then applied to clinical MSOT images from Crohn's disease patients and healthy controls.
Results
The phasor method facilitated the identification of artifacts and spectral anomalies, significantly improving interpretability of MSOT data. It also outperformed conventional unmixing algorithms in terms of processing speed, making it suitable for real-time application. In clinical datasets, the method revealed distinct spectral patterns between Crohn's disease patients and healthy individuals, highlighting its ability to detect changes and differences in biologically complex systems.
Conclusions
This work establishes the phasor approach as a powerful tool for MSOT spectral unmixing, offering both speed and clarity in data interpretation. Its real-time capability and diagnostic potential support broader clinical adoption of MSOT for noninvasive disease characterization.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


