裁剪任务算法以解决在多机构数据集上训练的模型中的偏差。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
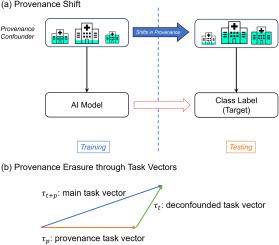
目的:多机构数据集广泛应用于临床数据的机器学习,以增加数据集规模并提高泛化能力。然而,特别是深度学习模型可能会学会识别数据元素的来源,从而导致有偏见的预测。例如,对来自不同数据源的COVID-19阳性和阴性胸片进行图像识别训练的深度学习模型可以响应来源指标(例如,每个机构特定实践的肺区域以外的放射学注释),而不是病理学,在其训练数据之外泛化性差。这种偏差被称为来源混淆,在自然语言处理(NLP)中是一个值得关注的问题,因为来源指标(例如,特定机构的章节标题,或特定地区的方言)在语言数据中普遍存在。先前解决这种偏见的工作主要集中在统计方法上,而没有为NLP的深度学习模型提供解决方案。方法:最近在表示学习方面的工作表明,将训练过的深度网络的权重表示为任务向量,允许它们的算术组合来控制模型对期望行为的能力。在这项工作中,我们评估了用这样的任务算法降低模型区分贡献站点的能力可以减轻来源混淆的程度。为此,我们提出了两种模型无关的方法,即来源效应约简的任务算法(TAPER)和优势对齐极化来源效应约简(DAPPER),将任务向量方法扩展到一个新的问题领域。结果:对三个数据集的评估表明,使用任务向量方法的RoBERTa和Llama-2模型对来源混淆的鲁棒性有所提高,在分布转移的极端情况下性能有所提高。结论:这项工作强调了调整来源混淆的重要性,特别是在极端情况下的转移。在使用深度学习模型时,DAPPER和锥度显示出减轻这种偏差的效率。它们提供了一种新的缓解来源混淆的策略,广泛适用于解决复合临床数据集中的其他偏倚来源。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Tailoring task arithmetic to address bias in models trained on multi-institutional datasets
Objective:
Multi-institutional datasets are widely used for machine learning from clinical data, to increase dataset size and improve generalization. However, deep learning models in particular may learn to recognize the source of a data element, leading to biased predictions. For example, deep learning models for image recognition trained on chest radiographs with COVID-19 positive and negative examples drawn from different data sources can respond to indicators of provenance (e.g., radiological annotations outside the lung area per institution-specific practices) rather than pathology, generalizing poorly beyond their training data. Bias of this sort, called confounding by provenance, is of concern in natural language processing (NLP) because provenance indicators (e.g., institution-specific section headers, or region-specific dialects) are pervasive in language data. Prior work on addressing such bias has focused on statistical methods, without providing a solution for deep learning models for NLP.
Methods:
Recent work in representation learning has shown that representing the weights of a trained deep network as task vectors allows for their arithmetic composition to govern model capabilities towards desired behaviors. In this work, we evaluate the extent to which reducing a model’s ability to distinguish between contributing sites with such task arithmetic can mitigate confounding by provenance. To do so, we propose two model-agnostic methods, Task Arithmetic for Provenance Effect Reduction (TAPER) and Dominance-Aligned Polarized Provenance Effect Reduction (DAPPER), extending the task vectors approach to a novel problem domain.
Results:
Evaluation on three datasets shows improved robustness to confounding by provenance for both RoBERTa and Llama-2 models with the task vector approach, with improved performance at the extremes of distribution shift.
Conclusion:
This work emphasizes the importance of adjusting for confounding by provenance, especially in extreme cases of the shift. In use of deep learning models, DAPPER and TAPER show efficiency in mitigating such bias. They provide a novel mitigation strategy for confounding by provenance, with broad applicability to address other sources of bias in composite clinical data sets. Source code is available within the DeconDTN toolkit: https://github.com/LinguisticAnomalies/DeconDTN-toolkit
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


