GatorCLR:使用自我监督对比图表示对电子健康记录中的患者结果进行个性化预测
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:近年来,人们对大量电子健康记录(EHR)数据的分析越来越感兴趣。患者预后预测是电子病历分析的一个主要研究领域,其重点是使用结构化数据类型(如从纵向电子病历数据收集的诊断、药物和程序)预测患者未来的健康状况。我们研究和设计了自监督学习(SSL)范式,从纵向电子病历数据中学习高质量的表示,旨在有效地捕获纵向关系和模式,以改进患者预后预测。方法:我们提出了一个名为GatorCLR的端到端、新颖且健壮的模型,该模型与对比SSL范例保持一致。GatorCLR将基于图分析的患者建模集成到纵向电子病历数据中,生成代表患者、患者关系和相似度的节点和边的图表示。我们的GatorCLR进一步采用了两层增强技术,该技术从图表示生成一致的、保持身份的增强。结果:我们使用真实世界的电子病历数据集评估我们的方法。实验结果表明,我们的GatorCLR在多个临床任务和数据集上提供了有意义和稳健的性能,并提供了模型决策的透明度。结论:所提出的方法为开发具有纵向EHR数据的基础模型迈出了重要的一步,该模型能够做出明智的预测,并适应各种下游用例和任务。因此,这项研究应该对希望利用纵向电子病历数据进行预测分析的从业者有价值。本文章由计算机程序翻译,如有差异,请以英文原文为准。
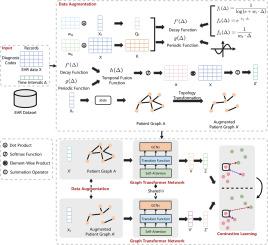
GatorCLR: Personalized predictions of patient outcomes on electronic health records using self-supervised contrastive graph representation
Objective:
Recently, there has been growing interest in analyzing large amounts of Electronic Health Record (EHR) data. Patient outcome prediction is a major area of interest in EHR analysis that focuses on predicting the future health status of patients using structured data types, such as diagnoses, medications, and procedures collected from longitudinal EHR data. We investigate and design self-supervised learning (SSL) paradigms to learn high-quality representations from longitudinal EHR data, aiming to effectively capture longitudinal relationships and patterns for improved patient outcome predictions.
Methods:
We propose an end-to-end, novel, and robust model called GatorCLR that aligns with the contrastive SSL paradigm. GatorCLR incorporates graph analysis-based patient modeling into longitudinal EHR data, generating graph representations of nodes and edges representing patients, their relationships, and similarities. A two-layer augmentation technique is further incorporated in our GatorCLR that generates consistent, identity-preserving augmentations from graph representations.
Results:
We evaluate our approach using real-world EHR datasets. Experimental results indicate that our GatorCLR delivers meaningful and robust performance across multiple clinical tasks and datasets and provides transparency of the model decisions.
Conclusion:
The proposed approach presents a significant step toward developing a foundation model with longitudinal EHR data, capable of making informed predictions and adaptable to various downstream use cases and tasks. This study should, therefore, be of value to practitioners wishing to leverage longitudinal EHR data for predictive analytics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


