通过生成建模的生物医学文本规范化。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
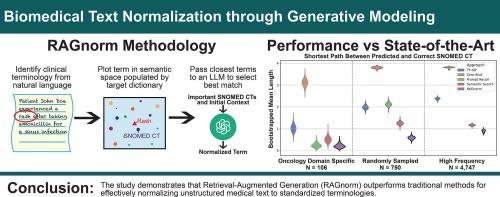
目的:电子健康档案(EHR)数据中有很大一部分是由非结构化的医学语言文本组成的。这种文本的格式通常是灵活的和不一致的,使得它具有挑战性,用于预测建模,临床决策支持和数据挖掘。大型语言模型(llm)理解上下文和语义变化的能力使它们成为标准化医学文本的有前途的工具。在这项研究中,我们开发并评估了使用大语言模型构建的临床文本规范化管道。方法:我们实现了四种基于llm的归一化策略(零概率召回、提示召回、语义搜索和基于检索增强生成的归一化[RAGnorm])和一种基于TF-IDF的字符串匹配的基线方法。我们评估了三个snomed -映射条件术语数据集的性能:[1]是肿瘤学特定数据集,[2]是机构医疗条件的代表性样本,[3]是来自我们机构的常见条件代码数据集(>1000次使用)。我们通过记录预测和真实SNOMED CT项之间的平均最短路径长度来测量性能。此外,我们根据TAC 2017药物标签注释对我们的模型进行了基准测试,该注释将术语标准化到医学词典监管活动(MedDRA)首选术语。结果:我们发现RAGnorm在每个数据集中都是最有效的,对于特定领域的数据集实现了0.21的平均最短路径长度,对于采样数据集实现了0.58,对于top terms数据集实现了0.90。在TAC2017会议的任务4中,它取得了88.01的微F1分数,在不查看提供的训练数据的情况下超越了所有其他模型。结论:我们发现以检索为中心的方法克服了传统LLM对这项任务的限制。生物医学自由文本规范化需要进一步探索RAGnorm及相关检索技术。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Biomedical text normalization through generative modeling
Objective
A large proportion of electronic health record (EHR) data consists of unstructured medical language text. The formatting of this text is often flexible and inconsistent, making it challenging to use for predictive modeling, clinical decision support, and data mining. Large language models’ (LLMs) ability to understand context and semantic variations makes them promising tools for standardizing medical text. In this study, we develop and assess clinical text normalization pipelines built using large-language models.
Methods
We implemented four LLM-based normalization strategies (Zero-Shot Recall, Prompt Recall, Semantic Search, and Retrieval-Augmented Generation based normalization [RAGnorm]) and one baseline approach using TF-IDF based String Matching. We evaluated performance across three datasets of SNOMED-mapped condition terms: [1] an oncology-specific dataset, [2] a representative sample of institutional medical conditions, and [3] a dataset of commonly occurring condition codes (>1000 uses) from our institution. We measured performance by recording the mean shortest path length between predicted and true SNOMED CT terms. Additionally, we benchmarked our models against the TAC 2017 drug label annotations, which normalizes terms to the Medical Dictionary for Regulatory Activities (MedDRA) Preferred Terms.
Results
We found that RAGnorm was the most effective throughout each dataset, achieving a mean shortest path length of 0.21 for the domain-specific dataset, 0.58 for the sampled dataset, and 0.90 for the top terms dataset. It achieved a micro F1 score of 88.01 on task 4 of the TAC2017 conference, surpassing all other models without viewing the provided training data.
Conclusion
We find that retrieval-focused approaches overcome traditional LLM limitations for this task. RAGnorm and related retrieval techniques should be explored further for the normalization of biomedical free text.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


