InstaNovo能够在大规模蛋白质组学实验中进行扩散驱动的从头肽测序
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
基于质谱的蛋白质组学侧重于识别产生串联质谱的肽段。传统方法依赖于蛋白质数据库,但在某些情况下往往受到限制或不适用。从头肽测序法在没有事先信息的情况下将肽序列分配到光谱上,对各种生物应用都很有价值;然而,由于缺乏准确性,它的应用仍然具有挑战性。在这里,我们介绍了将碎片离子峰转化为肽序列的转换器模型--InstaNovo。我们证明了 InstaNovo 优于最先进的方法,并展示了它在多个应用中的实用性。我们还介绍了 InstaNovo+,这是一种扩散模型,通过迭代完善预测序列来提高性能。利用这些模型,我们提高了治疗测序的覆盖率,发现了新的多肽,并在不同的数据集中检测到了未报告的生物体,从而扩大了蛋白质组学搜索的范围和检测率。我们的模型为直接蛋白质测序、免疫肽组学和黑暗蛋白质组探索等领域带来了机遇。本文章由计算机程序翻译,如有差异,请以英文原文为准。
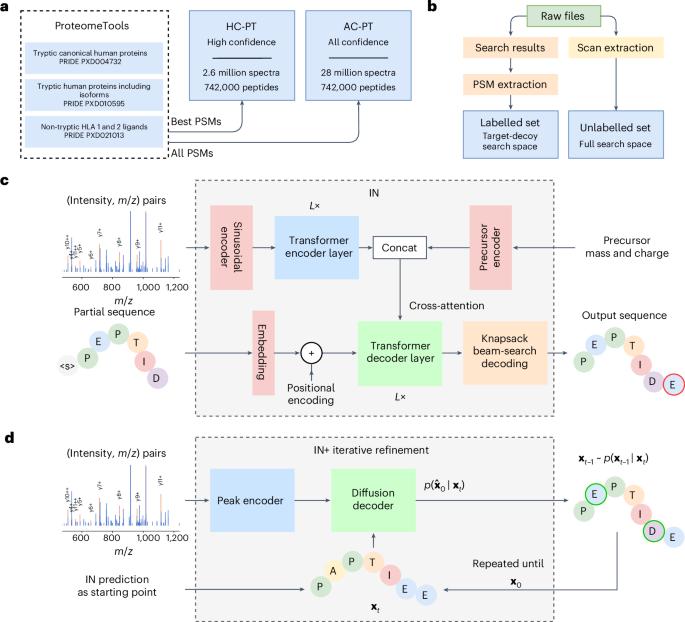

InstaNovo enables diffusion-powered de novo peptide sequencing in large-scale proteomics experiments
Mass spectrometry-based proteomics focuses on identifying the peptide that generates a tandem mass spectrum. Traditional methods rely on protein databases but are often limited or inapplicable in certain contexts. De novo peptide sequencing, which assigns peptide sequences to spectra without prior information, is valuable for diverse biological applications; however, owing to a lack of accuracy, it remains challenging to apply. Here we introduce InstaNovo, a transformer model that translates fragment ion peaks into peptide sequences. We demonstrate that InstaNovo outperforms state-of-the-art methods and showcase its utility in several applications. We also introduce InstaNovo+, a diffusion model that improves performance through iterative refinement of predicted sequences. Using these models, we achieve improved therapeutic sequencing coverage, discover novel peptides and detect unreported organisms in diverse datasets, thereby expanding the scope and detection rate of proteomics searches. Our models unlock opportunities across domains such as direct protein sequencing, immunopeptidomics and exploration of the dark proteome. InstaNovo, a transformer-based model, and InstaNovo+, a multinomial diffusion model, enhance de novo peptide sequencing, enabling discovery of novel peptides, improved therapeutics sequencing coverage and detection of unreported organisms in proteomics studies
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


