一个文本引导的蛋白质设计框架
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
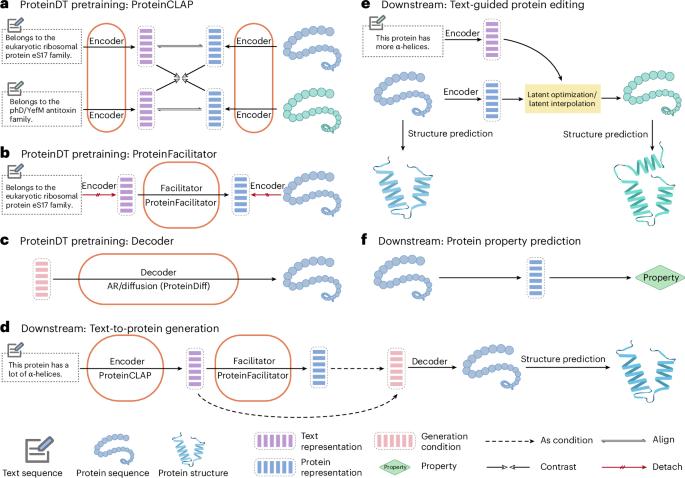
目前人工智能辅助蛋白质设计主要利用蛋白质序列和结构信息。与此同时,人类以文本形式整理了大量描述蛋白质高级功能的知识,但这些文本数据的结合是否有助于蛋白质设计任务尚未得到探索。为了弥补这一差距,我们提出了ProteinDT,这是一个利用文本描述进行蛋白质设计的多模态框架。ProteinDT由三个连续的步骤组成:ProteinCLAP,它使两种模式的表示保持一致,一个促进者从文本模式生成蛋白质表示,一个解码器从表示中创建蛋白质序列。为了训练ProteinDT,我们构建了一个大型数据集SwissProtCLAP,其中包含441,000个文本和蛋白质对。我们定量验证了ProteinDT在三个具有挑战性的任务上的有效性:(1)文本引导的蛋白质生成准确率超过90%;(2) 12个零射击文本引导蛋白编辑任务的最佳命中率;(3)在6个蛋白质特性预测指标中的4个指标上表现优异。本文章由计算机程序翻译,如有差异,请以英文原文为准。

A text-guided protein design framework
Current AI-assisted protein design utilizes mainly protein sequential and structural information. Meanwhile, there exists tremendous knowledge curated by humans in text format describing proteins’ high-level functionalities, yet whether the incorporation of such text data can help in protein design tasks has not been explored. To bridge this gap, we propose ProteinDT, a multimodal framework that leverages textual descriptions for protein design. ProteinDT consists of three consecutive steps: ProteinCLAP, which aligns the representation of two modalities, a facilitator that generates the protein representation from the text modality and a decoder that creates the protein sequences from the representation. To train ProteinDT, we construct a large dataset, SwissProtCLAP, with 441,000 text and protein pairs. We quantitatively verify the effectiveness of ProteinDT on three challenging tasks: (1) over 90% accuracy for text-guided protein generation; (2) best hit ratio on 12 zero-shot text-guided protein editing tasks; (3) superior performance on four out of six protein property prediction benchmarks. Shengchao Liu et al. present ProteinDT, a deep learning approach that can incorporate domain knowledge from textual descriptions into protein representation on a large scale.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


