一个全面的裂糖菌的物理转录因子与蛋白质和染色质相互作用图谱
IF 14.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
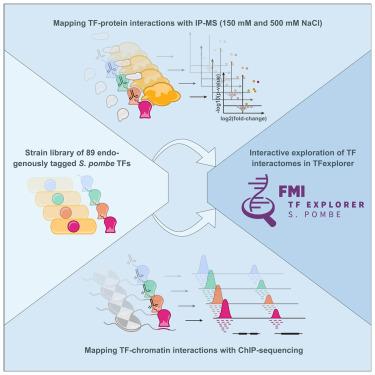
转录因子(tf)是基因表达的关键调控因子,但其许多靶点和作用方式尚不清楚。在Schizosaccharomyces pombe中,三分之一的TFs完全同源预测,很少有实验验证。我们创建了一个包含89个内源性标记的S. pombe tf的综合文库,使用免疫沉淀-质谱法和染色质免疫沉淀测序绘制了它们的蛋白质和染色质相互作用。我们的研究发现了一半TFs的蛋白质相互作用,其中超过四分之一可能形成稳定的复合物。我们在2027个独特的基因组区域中发现了大多数TF的dna结合位点,揭示了38个TF的基元,并揭示了一个广泛的TF交叉和自动调节的复杂网络。对最大的TF家族的表征揭示了保守的DNA序列偏好,但不同的结合模式,并鉴定了与核周基因定位相关的抑制异源二聚体Ntu1/Ntu2。我们的TFexplorer网络工具可以交互式访问所有数据,提供对TF相互作用和具有广泛生物学相关性的调节机制的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A comprehensive Schizosaccharomyces pombe atlas of physical transcription factor interactions with proteins and chromatin
Transcription factors (TFs) are key regulators of gene expression, yet many of their targets and modes of action remain unknown. In Schizosaccharomyces pombe, one-third of TFs are solely homology predicted, with few experimentally validated. We created a comprehensive library of 89 endogenously tagged S. pombe TFs, mapping their protein and chromatin interactions using immunoprecipitation-mass spectrometry and chromatin immunoprecipitation sequencing. Our study identified protein interactors for half the TFs, with over a quarter potentially forming stable complexes. We discovered DNA-binding sites for most TFs across 2,027 unique genomic regions, revealing motifs for 38 TFs and uncovering a complex network of extensive TF cross- and autoregulation. Characterization of the largest TF family revealed conserved DNA sequence preferences but diverse binding patterns and identified a repressive heterodimer, Ntu1/Ntu2, linked to perinuclear gene localization. Our TFexplorer webtool makes all data interactively accessible, offering insights into TF interactions and regulatory mechanisms with broad biological relevance.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


