蛋白袋包深导联优化及其在LTK蛋白强效选择性配体设计中的应用
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
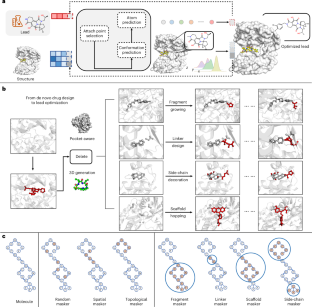
通过系统修饰来优化有前途的候选药物的化学结构以提高其效力和理化性质是药物发现管道中的重要步骤。与完善的从头生成方案相比,专门为铅优化量身定制的计算方法在很大程度上仍未得到充分开发。先前的模型通常局限于解决特定的子任务,例如生成二维分子结构,而忽略了三维空间中关键的蛋白质-配体相互作用。为了克服这些挑战,我们提出了Delete (Deep lead optimization in protein pocket),这是一种结合生成人工智能和基于结构的方法的一站式引线优化解决方案。该模型可以通过统一的删除(屏蔽)策略处理导联优化的所有子任务,并通过等变网络设计解释复杂的口袋配体相互作用。单个子任务的统计评估和回顾性研究表明,Delete具有出色的能力,可以使用给定的片段或原子来制造具有优越蛋白质结合能和合理药物相似性的分子。随后,我们利用Delete来设计针对先前鉴定的LTK蛋白的抑制剂。在Delete设计的配体中,CA-B-1通过体外和体内实验成功验证为有效的(1.36 nM)选择性抑制剂。本研究成功实现了基于结构的先导物优化模型Delete,用于快速可控的合理药物设计。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Deep lead optimization enveloped in protein pocket and its application in designing potent and selective ligands targeting LTK protein
Optimizing the chemical structure of promising drug candidates through systematic modifications to improve potency and physiochemical properties is a vital step in the drug discovery pipeline. In contrast to the well-established de novo generation schemes, computational methods specifically tailored for lead optimization remain largely underexplored. Prior models are often limited to addressing specific subtasks, such as generating two-dimensional molecular structures, while neglecting crucial protein–ligand interactions in three-dimensional space. To overcome these challenges, we propose Delete (Deep lead optimization enveloped in protein pocket), a one-stop solution for lead optimization by combining generative artificial intelligence and structure-based approaches. Our model can handle all subtasks of lead optimization through a unified deleting (masking) strategy, and it accounts for intricate pocket–ligand interactions through an equivariant network design. Statistical assessments and retrospective studies across individual subtasks demonstrate that Delete has an outstanding ability to craft molecules with superior protein-binding energy and reasonable drug-likeness using given fragments or atoms. Subsequently, we utilize Delete to design inhibitors targeting the previously identified LTK protein. Among the ligands designed by Delete, CA-B-1 is successfully validated as a potent (1.36 nM) and selective inhibitor by in vitro and in vivo experiments. This work represents a successful implementation of the powerful structure-based lead optimization model, Delete, for rapid and controllable rational drug design. Chen et al. present a deep learning-based lead optimization model that combines generative artificial intelligence with structure-based approaches. The method is successfully applied to the design of drug-like molecules targeting the recently identified LTK protein target with high potency and selectivity.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


