从人类粪便中摄取食物的宏基因组估计
IF 18.9
1区 医学
Q1 ENDOCRINOLOGY & METABOLISM
引用次数: 0
摘要
饮食摄入与肠道菌群组成、人体代谢和几乎所有主要慢性疾病的发病率密切相关。饮食和营养摄入通常采用自我报告方法进行评估,包括饮食问卷调查和食物记录,这些方法存在报告偏差,需要研究参与者严格遵守。在这里,我们提出了膳食摄入宏基因组估计(MEDI):一种量化人类粪便宏基因组中食物来源DNA的方法。我们表明,在粪便衍生的宏基因组数据中可以可靠地检测到含有dna的食物成分,即使丰度很低(超过10个reads)。我们展示了MEDI饮食摄入概况如何转化为营养摄入的详细代谢表征。MEDI确定了婴儿开始摄入固体食物的情况,与成年人的食物频率问卷调查结果一致,并与两项对照喂养研究中的食物和营养摄入量一致。最后,我们在一个没有饮食记录的大型临床队列中确定了与代谢综合征相关的特定饮食特征,为详细跟踪个人特定的、与健康相关的饮食模式提供了概念证明,而无需进行问卷调查。本文章由计算机程序翻译,如有差异,请以英文原文为准。

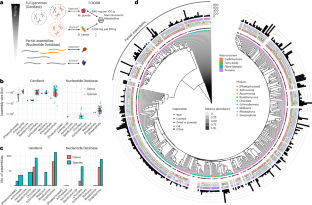
Metagenomic estimation of dietary intake from human stool
Dietary intake is tightly coupled to gut microbiota composition, human metabolism and the incidence of virtually all major chronic diseases. Dietary and nutrient intake are usually assessed using self-reporting methods, including dietary questionnaires and food records, which suffer from reporting biases and require strong compliance from study participants. Here, we present Metagenomic Estimation of Dietary Intake (MEDI): a method for quantifying food-derived DNA in human faecal metagenomes. We show that DNA-containing food components can be reliably detected in stool-derived metagenomic data, even when present at low abundances (more than ten reads). We show how MEDI dietary intake profiles can be converted into detailed metabolic representations of nutrient intake. MEDI identifies the onset of solid food consumption in infants, shows significant agreement with food frequency questionnaire responses in an adult population and shows agreement with food and nutrient intake in two controlled-feeding studies. Finally, we identify specific dietary features associated with metabolic syndrome in a large clinical cohort without dietary records, providing a proof-of-concept for detailed tracking of individual-specific, health-relevant dietary patterns without the need for questionnaires. Diener et al. present a method that allows the estimation of dietary intake from human stool by detecting food-derived DNA in faecal metagenomes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature metabolism
ENDOCRINOLOGY & METABOLISM-
CiteScore
27.50
自引率
2.40%
发文量
170
期刊介绍:
Nature Metabolism is a peer-reviewed scientific journal that covers a broad range of topics in metabolism research. It aims to advance the understanding of metabolic and homeostatic processes at a cellular and physiological level. The journal publishes research from various fields, including fundamental cell biology, basic biomedical and translational research, and integrative physiology. It focuses on how cellular metabolism affects cellular function, the physiology and homeostasis of organs and tissues, and the regulation of organismal energy homeostasis. It also investigates the molecular pathophysiology of metabolic diseases such as diabetes and obesity, as well as their treatment. Nature Metabolism follows the standards of other Nature-branded journals, with a dedicated team of professional editors, rigorous peer-review process, high standards of copy-editing and production, swift publication, and editorial independence. The journal has a high impact factor, has a certain influence in the international area, and is deeply concerned and cited by the majority of scholars.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


