PhageDPO:一个基于机器学习的计算框架,用于识别噬菌体解聚合酶
IF 6.3
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
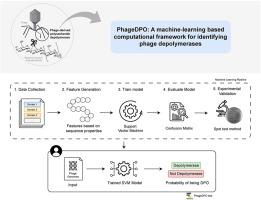
噬菌体(噬菌体)是地球上最具优势和遗传多样性的生物实体。噬菌体是一种感染细菌并编码多种具有潜在生物技术应用价值的蛋白质的病毒。然而,大多数噬菌体编码蛋白在功能上仍未被表征。解聚合酶(DPOs)是一种降解外部多糖结构的酶,从基础研究的角度和生物技术应用的角度来看,它已经引起了越来越多的兴趣,以控制细菌病原体。尽管预测噬菌体基因组中DPOs的鉴定工具越来越多,但我们介绍了PhageDPO作为一种强大而可靠的解决方案。PhageDPO在一个综合数据集上进行训练,该数据集包括与七个特定dpo相关结构域相关的序列,并在文献中验证了dpo。训练支持向量机(SVM)模型的测试准确率为96%,召回率为97%,精度为94%,f1分数为96%,证明了其预测噬菌体基因组DPOs的能力。利用文献报道的病例和本研究新生成的数据进一步验证了模型,提高了模型的性能。除了预测性能之外,PhageDPO还提供了一个用户友好的界面,加上强大的性能,使其与其他具有图形界面的工具相比更易于访问和有效。本文章由计算机程序翻译,如有差异,请以英文原文为准。
PhageDPO: A machine-learning based computational framework for identifying phage depolymerases
Bacteriophages (phages) are the most predominant and genetically diverse biological entities on Earth. Phages are viruses that infect bacteria and encode numerous proteins with potential biotechnological application. However, most phage-encoded proteins remain functionally uncharacterized. Depolymerases (DPOs) in particular, enzymes that degrade external polysaccharide structures, have garnered increasing interest from both fundamental research standpoint and for biotechnological applications to control bacterial pathogens. Despite the proliferation of identification tools for predicting DPOs in phage genomes, we introduced PhageDPO as a robust and reliable solution. PhageDPO is trained on a comprehensive dataset that includes sequences related to seven specific DPO-related domains, completed with DPOs validated in the literature. Training a Support Vector Machine (SVM) model resulted in a test accuracy of 96 %, a recall of 97 %, a precision of 94 % and a F1-score of 96 %, demonstrating its capability in predicting DPOs in phage genomes. The model was further validated using both cases reported in the literature and newly generated data for this study, enhancing its performance. Beyond its predictive performance, PhageDPO distinguishes itself by offering a user-friendly interface coupled with robust performance, making it more accessible and effective compared to other tools with graphical interfaces.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


