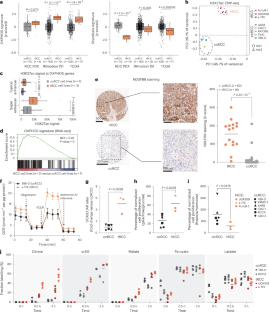
在易位性肾细胞癌中,致癌的TFE3融合驱动OXPHOS并赋予代谢脆弱性
IF 18.9
1区 医学
Q1 ENDOCRINOLOGY & METABOLISM
引用次数: 0
摘要
易位肾细胞癌(tRCC)是由 TFE3 基因融合驱动的一种侵袭性肾癌亚型,其下游机制特征不清。在这里,我们报告了 TFE3 融合基因通过转录重新连接 tRCC,使其趋向氧化磷酸化(OXPHOS),这与大多数其他肾癌的高糖酵解性质形成了鲜明对比。依赖这种 TFE3 融合驱动的 OXPHOS 方案会使 tRCC 易受 NADH 还原压力的影响,而 NADH 还原压力是一种由还原当量失衡引起的代谢压力。基因组规模的CRISPR筛选确定了与这种代谢状态相关的tRCC选择性脆弱性,包括EGLN1,它能羟化HIF-1α并使其成为蛋白水解的靶标。抑制 EGLN1 会稳定 HIF-1α,促进代谢重编程,使其脱离 OXPHOS,从而影响 tRCC 细胞的生长,这也是依赖 OXPHOS 的 tRCC 细胞的一个弱点。我们的研究确定了 tRCC 依赖于由 TFE3 融合驱动的以线粒体为中心的代谢程序,并将 EGLN1 抑制作为治疗这种癌症的一种策略。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Oncogenic TFE3 fusions drive OXPHOS and confer metabolic vulnerabilities in translocation renal cell carcinoma
Translocation renal cell carcinoma (tRCC) is an aggressive subtype of kidney cancer driven by TFE3 gene fusions, which act via poorly characterized downstream mechanisms. Here we report that TFE3 fusions transcriptionally rewire tRCCs toward oxidative phosphorylation (OXPHOS), contrasting with the highly glycolytic nature of most other renal cancers. Reliance on this TFE3 fusion-driven OXPHOS programme renders tRCCs vulnerable to NADH reductive stress, a metabolic stress induced by an imbalance of reducing equivalents. Genome-scale CRISPR screening identifies tRCC-selective vulnerabilities linked to this metabolic state, including EGLN1, which hydroxylates HIF-1α and targets it for proteolysis. Inhibition of EGLN1 compromises tRCC cell growth by stabilizing HIF-1α and promoting metabolic reprogramming away from OXPHOS, thus representing a vulnerability for OXPHOS-dependent tRCC cells. Our study defines tRCC as being dependent on a mitochondria-centred metabolic programme driven by TFE3 fusions and nominates EGLN1 inhibition as a therapeutic strategy in this cancer. Li et al. show that the TFE3 driver fusion transcriptionally upregulates an OXPHOS programme in translocation renal cell carcinoma that renders this cancer vulnerable to reductive stress, induced by imbalance of reducing equivalents.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature metabolism
ENDOCRINOLOGY & METABOLISM-
CiteScore
27.50
自引率
2.40%
发文量
170
期刊介绍:
Nature Metabolism is a peer-reviewed scientific journal that covers a broad range of topics in metabolism research. It aims to advance the understanding of metabolic and homeostatic processes at a cellular and physiological level. The journal publishes research from various fields, including fundamental cell biology, basic biomedical and translational research, and integrative physiology. It focuses on how cellular metabolism affects cellular function, the physiology and homeostasis of organs and tissues, and the regulation of organismal energy homeostasis. It also investigates the molecular pathophysiology of metabolic diseases such as diabetes and obesity, as well as their treatment. Nature Metabolism follows the standards of other Nature-branded journals, with a dedicated team of professional editors, rigorous peer-review process, high standards of copy-editing and production, swift publication, and editorial independence. The journal has a high impact factor, has a certain influence in the international area, and is deeply concerned and cited by the majority of scholars.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


