深度学习增强了外源病原体中HLA i类CD8+ T细胞表位的预测
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
CD8+ T细胞表位的精确计算机测定将极大地促进基于T细胞的疫苗开发,但目前的预测模型并不可靠。在这里,受最近成功将机器学习应用于复杂生物学的激励,我们整理了651,237个独特的人类白细胞抗原I类(HLA-I)配体的数据集,并开发了MUNIS,这是一种深度学习模型,用于识别HLA-I等位基因呈现的肽。与现有模型相比,MUNIS在预测肽呈递和CD8+ T细胞表位免疫优势等级方面表现出更高的性能。此外,将MUNIS应用于eb病毒蛋白,成功鉴定了已建立的和新的hla - 1表位,并通过体外hla - 1肽稳定性和T细胞免疫原性实验验证了这些表位。MUNIS在免疫原性预测方面的表现与实验稳定性分析相当,这表明深度学习可以减轻实验负担并加速CD8+ T细胞表位的鉴定,从而快速开发T细胞疫苗。本文章由计算机程序翻译,如有差异,请以英文原文为准。

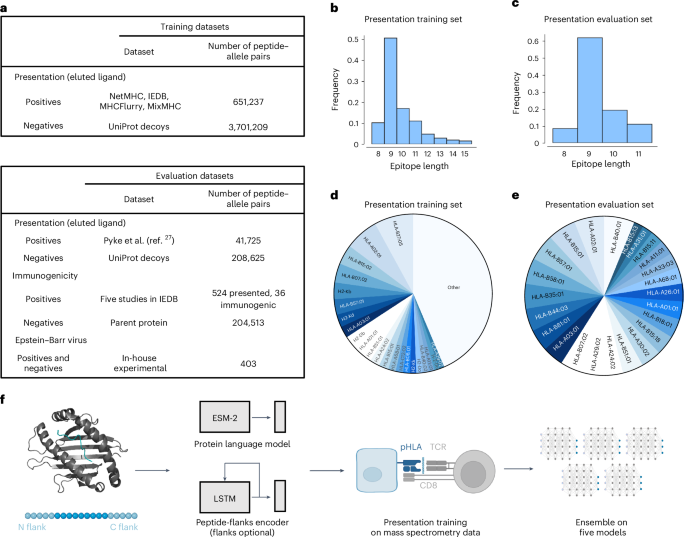
Deep learning enhances the prediction of HLA class I-presented CD8+ T cell epitopes in foreign pathogens
Accurate in silico determination of CD8+ T cell epitopes would greatly enhance T cell-based vaccine development, but current prediction models are not reliably successful. Here, motivated by recent successes applying machine learning to complex biology, we curated a dataset of 651,237 unique human leukocyte antigen class I (HLA-I) ligands and developed MUNIS, a deep learning model that identifies peptides presented by HLA-I alleles. MUNIS shows improved performance compared with existing models in predicting peptide presentation and CD8+ T cell epitope immunodominance hierarchies. Moreover, application of MUNIS to proteins from Epstein–Barr virus led to successful identification of both established and novel HLA-I epitopes which were experimentally validated by in vitro HLA-I-peptide stability and T cell immunogenicity assays. MUNIS performs comparably to an experimental stability assay in terms of immunogenicity prediction, suggesting that deep learning can reduce experimental burden and accelerate identification of CD8+ T cell epitopes for rapid T cell vaccine development. Accurate prediction of immunogenic CD8+ T cell epitopes would greatly accelerate T cell vaccine development. A new deep learning model, MUNIS, can rapidly identify HLA-binding, immunogenic and immunodominant peptides in foreign pathogens.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


