迈向高度敏感的基于深度学习的端到端串联质谱数据库搜索
IF 23.9
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
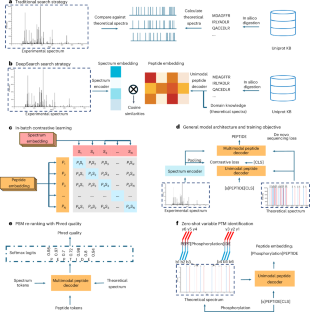
基于质谱的蛋白质组学中的肽鉴定对于理解蛋白质的功能和动力学至关重要。传统的数据库搜索方法虽然被广泛使用,但依赖于启发式评分函数,必须引入统计估计才能达到更高的识别率。本文介绍了一种基于深度学习的串联质谱端到端数据库搜索方法DeepSearch。DeepSearch在对比学习框架下利用改进的基于变压器的编码器-解码器架构。与依赖离子对离子匹配的传统方法不同,DeepSearch采用数据驱动的方法对肽谱匹配进行评分。DeepSearch还可以以零采样的方式分析翻译后的变化。我们证明了DeepSearch的评分方案表达了更少的偏差,并且不需要任何统计估计。我们在各种数据集上验证了DeepSearch的准确性和稳健性,包括来自具有不同蛋白质组成的物种的数据集和修改丰富的数据集。DeepSearch为串联质谱的数据库搜索方法提供了新的思路。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Towards highly sensitive deep learning-based end-to-end database search for tandem mass spectrometry
Peptide identification in mass spectrometry-based proteomics is crucial for understanding protein function and dynamics. Traditional database search methods, though widely used, rely on heuristic scoring functions, and statistical estimations must be introduced to achieve a higher identification rate. Here we introduce DeepSearch, a deep learning-based end-to-end database search method for tandem mass spectrometry. DeepSearch leverages a modified transformer-based encoder–decoder architecture under the contrastive learning framework. Unlike conventional methods, which rely on ion-to-ion matching, DeepSearch adopts a data-driven approach to score peptide–spectrum matches. DeepSearch can also profile variable post-translational modifications in a zero-shot manner. We show that DeepSearch’s scoring scheme expresses less bias and does not require any statistical estimation. We validate DeepSearch’s accuracy and robustness across various datasets, including those from species with diverse protein compositions and a modification-enriched dataset. DeepSearch sheds new light on database search methods in tandem mass spectrometry. Yu and Li present DeepSearch, a deep learning-based method for peptide identification in mass spectrometry, offering unbiased, data-driven scoring without statistical estimation. It accurately profiles post-translational modifications in a zero-shot manner.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


