使用紧凑特征集将非TCGA癌症样本分类为TCGA分子亚型
IF 48.8
1区 医学
Q1 CELL BIOLOGY
引用次数: 0
摘要
由癌症基因组图谱(TCGA)定义的分子亚型描述了癌症的潜在生物学特征,为患者的预后和治疗计划带来了希望。然而,大多数用于发现亚型的方法并不适合为来自其他研究或临床试验的新癌症标本分配亚型标签。在这里,我们通过应用五种不同的机器学习方法来解决这一障碍,这些方法来自8,791个TCGA肿瘤样本,包括来自26个不同癌症队列的106个亚型,以建立基于少量特征的模型,这些特征可以将新样本分类为先前定义的TCGA分子亚型,这是分子亚型在临床应用的一步。我们使用外部数据集验证选择的分类器。预测性能和分类器选择的特征可以深入了解不同的机器学习方法和基因组数据平台。对于每种癌症和数据类型,我们都将性能最好的模型的容器化版本作为公共资源提供。本文章由计算机程序翻译,如有差异,请以英文原文为准。
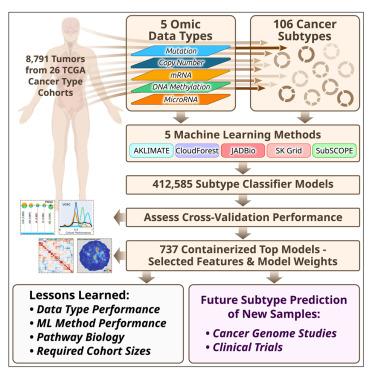
Classification of non-TCGA cancer samples to TCGA molecular subtypes using compact feature sets
Molecular subtypes, such as defined by The Cancer Genome Atlas (TCGA), delineate a cancer’s underlying biology, bringing hope to inform a patient’s prognosis and treatment plan. However, most approaches used in the discovery of subtypes are not suitable for assigning subtype labels to new cancer specimens from other studies or clinical trials. Here, we address this barrier by applying five different machine learning approaches to multi-omic data from 8,791 TCGA tumor samples comprising 106 subtypes from 26 different cancer cohorts to build models based upon small numbers of features that can classify new samples into previously defined TCGA molecular subtypes—a step toward molecular subtype application in the clinic. We validate select classifiers using external datasets. Predictive performance and classifier-selected features yield insight into the different machine-learning approaches and genomic data platforms. For each cancer and data type we provide containerized versions of the top-performing models as a public resource.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cancer Cell
医学-肿瘤学
CiteScore
55.20
自引率
1.20%
发文量
179
审稿时长
4-8 weeks
期刊介绍:
Cancer Cell is a journal that focuses on promoting major advances in cancer research and oncology. The primary criteria for considering manuscripts are as follows:
Major advances: Manuscripts should provide significant advancements in answering important questions related to naturally occurring cancers.
Translational research: The journal welcomes translational research, which involves the application of basic scientific findings to human health and clinical practice.
Clinical investigations: Cancer Cell is interested in publishing clinical investigations that contribute to establishing new paradigms in the treatment, diagnosis, or prevention of cancers.
Insights into cancer biology: The journal values clinical investigations that provide important insights into cancer biology beyond what has been revealed by preclinical studies.
Mechanism-based proof-of-principle studies: Cancer Cell encourages the publication of mechanism-based proof-of-principle clinical studies, which demonstrate the feasibility of a specific therapeutic approach or diagnostic test.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


