越南全绢波马属植物ITS及部分叶绿体DNA区数据集。
IF 1.4
Q3 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
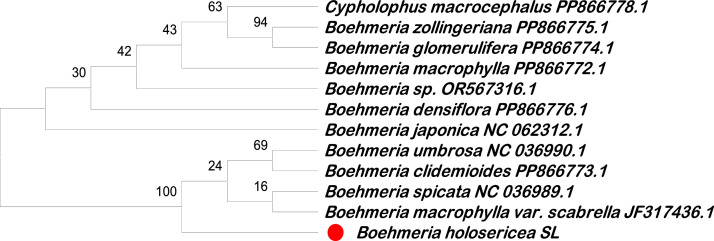
苧麻属的物种有可能成为天然药物,并具有工业纤维生产用途。该属的许多物种形态相似,很难区分,尤其是当它们的形态扭曲时。本数据集包括从苧麻基因组中分离出的几个 DNA 区域的序列信息,即 ITS(来自核基因组)、matK、trnL-trnF、trnH-psbA 和 rpoC1(来自叶绿体基因组),以及基于分离序列的系统发生分析结果。在基于 matK 基因序列的系统发生树上,B. holosericea 与 B. umbrosa、B. clidemioides、B. spicata 和 B. macrophylla 被归为一类,引导系数为 100%。在基于 trnH-psbA spacer 区域序列的系统发生树中,B. holosericea 与 B. clidemioides 被归为一类(bootstrap coefficient 为 96%)。在基于 rpoC1 基因序列的系统发生树中,B. holosericea 与 B. spicata 被归为一类(引导系数为 100%)。在基于 ITS 区域序列的系统发生树中,B. holosericea 与 B. macrophylla 被归为一类(bootstrap 系数为 73%);基于 trnL-trnF spacer 区域,B. holosericea 与 B. pilociuscula 被归为一类(bootstrap 系数为 16%)。叶绿体基因组中的两个基因 matK 和 rpoC1 以及 trnH-psbA 区域是潜在的 DNA 条形码候选者,可帮助鉴定全丝核菌的物种。该数据集首次报道了 ITS、matK、trnL-trnF、trnH-psbA 和 rpoC1 序列以及 B. holosericea 的系统发育。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Dataset on ITS and some chloroplast DNA regions of Boehmeria holosericea Blume in Vietnam
Species of the Boehmeria genus have the potential to be natural medicines and have industrial fibre production uses. Many species of this genus are morphologically similar and are difficult to distinguish, especially when their morphology is distorted. This dataset includes sequence information of several DNA regions isolated from the genome of Boehmeria holosericea, namely ITS (from the nuclear genome), matK, trnL-trnF, trnH-psbA, and rpoC1 (from the chloroplast genome) and phylogenetic analysis results based on the isolated sequences. On the phylogenetic tree based on the matK gene sequence, B. holosericea is grouped with B. umbrosa, B. clidemioides, B. spicata, and B. macrophylla with a bootstrap coefficient of 100%. In the phylogenetic tree based on the trnH-psbA spacer region sequences, B. holosericea was grouped with B. clidemioides (a bootstrap coefficient of 96%). In the phylogenetic tree based on the rpoC1 gene sequences, B. holosericea was grouped with B. spicata (a bootstrap coefficient of 100%). In the phylogenetic tree based on the ITS region sequences, B. holosericea was grouped with B. macrophylla (a bootstrap coefficient of 73%), and based on the trnL-trnF spacer region, B. holosericea was grouped with B. pilociuscula (a bootstrap coefficient of 16%). Two genes, matK and rpoC1 and the trnH-psbA region from the chloroplast genome, are potential DNA barcode candidates that could aid in the species identification of B. holosericea. This dataset the first report on the ITS, matK, trnL-trnF, trnH-psbA, and rpoC1 sequences and the phylogeny of B. holosericea.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Data in Brief
MULTIDISCIPLINARY SCIENCES-
CiteScore
3.10
自引率
0.00%
发文量
996
审稿时长
70 days
期刊介绍:
Data in Brief provides a way for researchers to easily share and reuse each other''s datasets by publishing data articles that: -Thoroughly describe your data, facilitating reproducibility. -Make your data, which is often buried in supplementary material, easier to find. -Increase traffic towards associated research articles and data, leading to more citations. -Open up doors for new collaborations. Because you never know what data will be useful to someone else, Data in Brief welcomes submissions that describe data from all research areas.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


