分布变化条件下用于生存分析的稳定考克斯回归
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
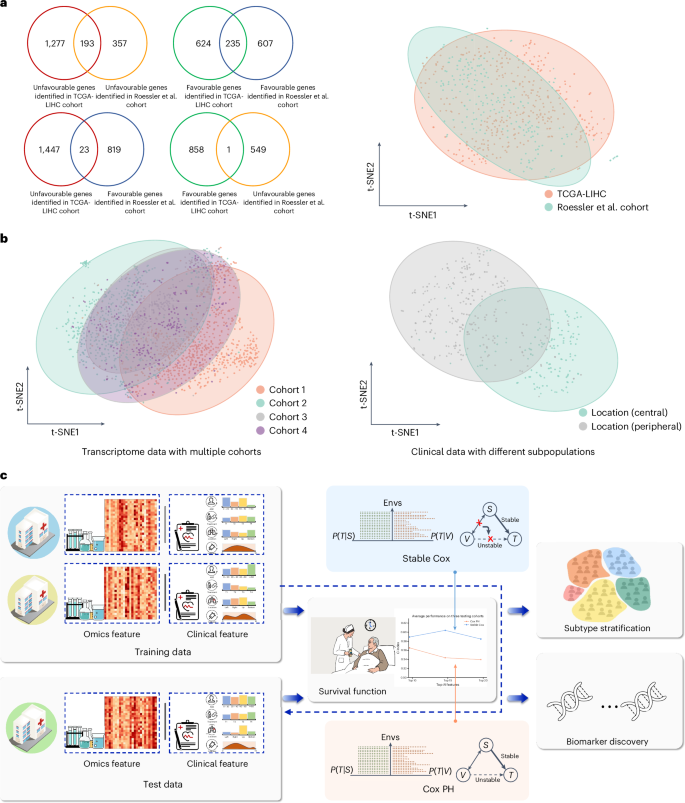
生存分析的目的是估计协变量对事件发生前预期时间的影响,这在生命科学和医疗保健等学科中被广泛应用,极大地影响决策并改善生存结果。现有的方法,通常假设类似的训练和测试分布,然而面对现实世界中不同数据源的挑战,产生不可预测的变化,破坏了它们的可靠性。这迫切需要生存分析方法应该利用不同群体的稳定特征进行预测,而不是依赖于虚假的相关性。为此,我们提出了一个具有理论保证的稳定Cox模型来识别稳定变量,该模型联合优化了独立性驱动的样本重赋权模块和加权Cox回归模型。通过对模拟组学和现实组学以及临床数据的广泛评估,稳定的Cox不仅在不同的独立测试集上表现出较强的泛化能力,而且可以通过鉴定的生物标志物面板显著地划分患者的亚型。本文章由计算机程序翻译,如有差异,请以英文原文为准。

Stable Cox regression for survival analysis under distribution shifts
Survival analysis aims to estimate the impact of covariates on the expected time until an event occurs, which is broadly utilized in disciplines such as life sciences and healthcare, substantially influencing decision-making and improving survival outcomes. Existing methods, usually assuming similar training and testing distributions, nevertheless face challenges with real-world varying data sources, creating unpredictable shifts that undermine their reliability. This urgently necessitates that survival analysis methods should utilize stable features across diverse cohorts for predictions, rather than relying on spurious correlations. To this end, we propose a stable Cox model with theoretical guarantees to identify stable variables, which jointly optimizes an independence-driven sample reweighting module and a weighted Cox regression model. Through extensive evaluation on simulated and real-world omics and clinical data, stable Cox not only shows strong generalization ability across diverse independent test sets but also stratifies the subtype of patients significantly with the identified biomarker panels. Survival prediction models used in healthcare usually assume that training and test data share a similar distribution, which is not true in real-world settings. Cui and colleagues develop a stable Cox regression model that can identify stable variables for predicting survival outcomes under distribution shifts.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


