表观转录组 rRNA 指纹图谱揭示了原发组织和肿瘤特异性特征
IF 16.6
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
哺乳动物核糖体RNA (rRNA)分子是高度丰富的RNA,有220多种修饰。先前的研究表明,一些rRNA修饰类型可以被动态调节;然而,哺乳动物rRNA修饰景观如何以及何时被重塑在很大程度上仍未被探索。在这里,我们采用直接RNA测序来绘制人类和小鼠rRNA表转录组在组织、发育阶段、细胞类型和疾病中的表转录组。我们的分析揭示了多个rRNA位点以组织和/或发育阶段特异性的方式进行差异修饰,包括以前未注释的修饰位点。我们证明,rRNA修饰模式可用于组织和细胞类型鉴定,我们在此称之为“表转录组指纹图谱”。然后,我们探索了来自肺癌患者的正常肿瘤匹配样本中的rRNA修饰模式,发现表转录组指纹技术仅从每个样本的250个reads中准确地将临床样本分为正常组和肿瘤组,证明了rRNA修饰作为诊断生物标志物的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
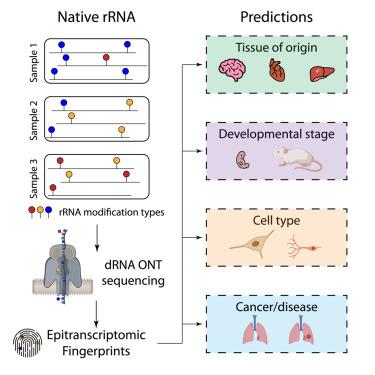
Epitranscriptomic rRNA fingerprinting reveals tissue-of-origin and tumor-specific signatures
Mammalian ribosomal RNA (rRNA) molecules are highly abundant RNAs, decorated with over 220 rRNA modifications. Previous works have shown that some rRNA modification types can be dynamically regulated; however, how and when the mammalian rRNA modification landscape is remodeled remains largely unexplored. Here, we employ direct RNA sequencing to chart the human and mouse rRNA epitranscriptome across tissues, developmental stages, cell types, and disease. Our analyses reveal multiple rRNA sites that are differentially modified in a tissue- and/or developmental stage-specific manner, including previously unannotated modified sites. We demonstrate that rRNA modification patterns can be used for tissue and cell-type identification, which we hereby term “epitranscriptomic fingerprinting.” We then explore rRNA modification patterns in normal-tumor matched samples from lung cancer patients, finding that epitranscriptomic fingerprinting accurately classifies clinical samples into normal and tumor groups from only 250 reads per sample, demonstrating the potential of rRNA modifications as diagnostic biomarkers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


