GABAA受体π通过g蛋白依赖途径形成刺激ERK的通道
IF 16.6
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
罕见的γ-氨基丁酸a型受体(GABAAR)亚基π (GABRP)在某些癌症中高度表达,它通过细胞外调节激酶(ERK)信号通路以一种未知的途径刺激肿瘤生长。为了阐明GABRP的信号机制,我们测定了在GABA存在和不存在的情况下嵌入天然纳米盘的GABRP的冷冻电镜(cryo-EM)结构。在结构上,GABRP同戊二聚体非常类似于异戊二聚GABAAR阴离子通道,在GABA结合后从封闭的“静息”状态转变为开放的“活性”状态。然而,功能分析显示GABRP的反应更像b型代谢受体。在GABA的生理浓度下,氯化物通量未被检测到。相反,GABRP激活了导致ERK信号传导的g蛋白偶联通路。电离性活性仅在超生理的GABA浓度下触发,有效地将其与GABRP的信号功能解耦。这些发现提供了GABRP的结构和功能蓝图,为GABRP阳性癌症中GABA生长信号的靶向抑制开辟了新的途径。本文章由计算机程序翻译,如有差异,请以英文原文为准。
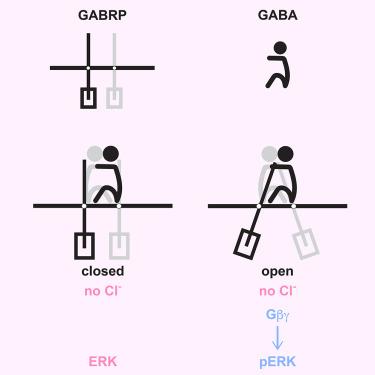
GABAA receptor π forms channels that stimulate ERK through a G-protein-dependent pathway
The rare γ-aminobutyric acid type-A receptor (GABAAR) subunit π (GABRP) is highly expressed in certain cancers, where it stimulates growth through extracellular-regulated kinase (ERK) signaling by an uncharacterized pathway. To elucidate GABRP’s signaling mechanism, we determined cryoelectron microscopy (cryo-EM) structures of GABRP embedded in native nanodiscs, both in the presence and absence of GABA. Structurally, GABRP homopentamers closely resemble heteropentameric GABAAR anion channels, transitioning from a closed “resting” state to an open “active” state upon GABA binding. However, functional assays reveal that GABRP responds more like a type-B metabotropic receptor. At physiological concentrations of GABA, chloride flux is not detected. Rather, GABRP activates a G-protein-coupled pathway leading to ERK signaling. Ionotropic activity is only triggered at supraphysiological GABA concentrations, effectively decoupling it from GABRP’s signaling functions. These findings provide a structural and functional blueprint for GABRP, opening new avenues for targeted inhibition of GABA growth signals in GABRP-positive cancers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


