通过详尽的高分辨率模板匹配重建人核糖体原位结构动力学
IF 16.6
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
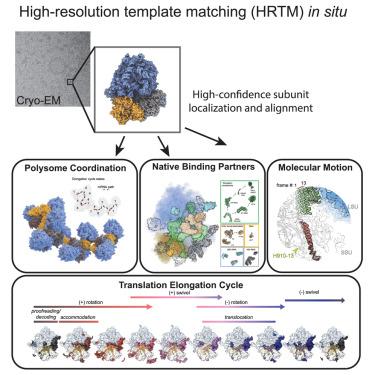
蛋白质合成是生命的核心,需要核糖体,核糖体通过一系列的结构转化催化氨基酸逐步添加到多肽链上。在这里,我们对直接冷冻固定的活细胞的冷冻电镜(cryo-EM)图像采用高分辨率模板匹配(HRTM),以获得覆盖整个延伸周期的一组核糖体构型,每种构型都以其天然丰度出现。HRTM的定位和取向精度以及检测小目标(~ 300 kDa)的能力使其能够沿着反应坐标排列这些构型,并沿着延伸周期重建任何构型的分子特征。通过将40个重建序列组合成3D电影,可视化循环的结构动力学很容易揭示组分和配体的运动,其中一些令人惊讶,如弹簧样的分子内运动,为链延伸过程中一些仍然神秘的步骤所涉及的分子机制提供线索。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structural dynamics of human ribosomes in situ reconstructed by exhaustive high-resolution template matching
Protein synthesis is central to life and requires the ribosome, which catalyzes the stepwise addition of amino acids to a polypeptide chain by undergoing a sequence of structural transformations. Here, we employed high-resolution template matching (HRTM) on cryoelectron microscopy (cryo-EM) images of directly cryofixed living cells to obtain a set of ribosomal configurations covering the entire elongation cycle, with each configuration occurring at its native abundance. HRTM’s position and orientation precision and ability to detect small targets (∼300 kDa) made it possible to order these configurations along the reaction coordinate and to reconstruct molecular features of any configuration along the elongation cycle. Visualizing the cycle’s structural dynamics by combining a sequence of >40 reconstructions into a 3D movie readily revealed component and ligand movements, some of them surprising, such as spring-like intramolecular motion, providing clues about the molecular mechanisms involved in some still mysterious steps during chain elongation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Cell
生物-生化与分子生物学
CiteScore
26.00
自引率
3.80%
发文量
389
审稿时长
1 months
期刊介绍:
Molecular Cell is a companion to Cell, the leading journal of biology and the highest-impact journal in the world. Launched in December 1997 and published monthly. Molecular Cell is dedicated to publishing cutting-edge research in molecular biology, focusing on fundamental cellular processes. The journal encompasses a wide range of topics, including DNA replication, recombination, and repair; Chromatin biology and genome organization; Transcription; RNA processing and decay; Non-coding RNA function; Translation; Protein folding, modification, and quality control; Signal transduction pathways; Cell cycle and checkpoints; Cell death; Autophagy; Metabolism.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


