吩噻嗪衍生物:合成、对接研究和抗菌活性
IF 4
2区 化学
Q2 CHEMISTRY, PHYSICAL
引用次数: 0
摘要
研究了5种吩噻嗪衍生物A、B、C、D和E对3种革兰氏阳性菌和3种革兰氏阴性菌以及7种真菌的抑菌效果。对收集到的数据进行分析,发现所有的衍生物都具有很强的抗菌性能。然而,它们作为抗真菌药物的效果相对较差,这可能与真菌的细胞结构有关。值得注意的是,在一些情况下,吩噻嗪衍生物的活性超过了参考药物青霉素10。此外,分子对接研究提供了这四种衍生物与靶酶PBP2a结合能力的额外证据。得到的对接分数范围为-11.27 ~ -15.90 Kcal/mol。根据药代动力学和ADME研究,所有吩噻嗪衍生物a - e以及青霉素10的生物利用度均高于零,范围在0.17 ~ 0.55之间。吩噻嗪衍生物A与参比药青霉素10相似,说明其生物活性最强。本文章由计算机程序翻译,如有差异,请以英文原文为准。
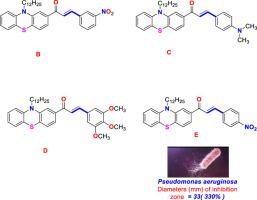
Phenothiazine derivatives: Synthesis, docking studies and antimicrobial activity
The antimicrobial effectiveness of five phenothiazine derivatives, namely A, B, C, D and E, was examined against three Gram-positive and three Gram-negative bacteria, as well as seven fungal species. Analysis of the gathered data revealed that all derivatives exhibited strong antibacterial properties. However, their performance as antifungal agents were comparatively less effective which may be attributed to cell structure of fungi. Notably, in several instances, the activity of the phenothiazine derivatives surpassed that of the reference drug penicillin 10. Furthermore, molecular docking studies provided additional evidence of the ability of these four derivatives to bind with the target enzyme PBP2a. The docking scores obtained ranged from -11.27 to -15.90 Kcal/mol. According to pharmacokinetics and ADME study, all phenothiazine derivatives A-E, as well as Penicillin 10, have a bioavailability higher than zero, ranging between 0.17 and 0.55. Phenothiazine derivative A is similar to the reference drug penicillin 10, indicating that it is the most bioactive.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Journal of Molecular Structure
化学-物理化学
CiteScore
7.10
自引率
15.80%
发文量
2384
审稿时长
45 days
期刊介绍:
The Journal of Molecular Structure is dedicated to the publication of full-length articles and review papers, providing important new structural information on all types of chemical species including:
• Stable and unstable molecules in all types of environments (vapour, molecular beam, liquid, solution, liquid crystal, solid state, matrix-isolated, surface-absorbed etc.)
• Chemical intermediates
• Molecules in excited states
• Biological molecules
• Polymers.
The methods used may include any combination of spectroscopic and non-spectroscopic techniques, for example:
• Infrared spectroscopy (mid, far, near)
• Raman spectroscopy and non-linear Raman methods (CARS, etc.)
• Electronic absorption spectroscopy
• Optical rotatory dispersion and circular dichroism
• Fluorescence and phosphorescence techniques
• Electron spectroscopies (PES, XPS), EXAFS, etc.
• Microwave spectroscopy
• Electron diffraction
• NMR and ESR spectroscopies
• Mössbauer spectroscopy
• X-ray crystallography
• Charge Density Analyses
• Computational Studies (supplementing experimental methods)
We encourage publications combining theoretical and experimental approaches. The structural insights gained by the studies should be correlated with the properties, activity and/ or reactivity of the molecule under investigation and the relevance of this molecule and its implications should be discussed.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


