一种识别多靶点JAK/STAT抑制剂作为抗癌药物的新型计算机方法
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
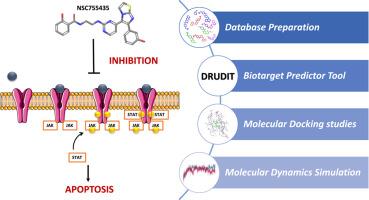
细胞凋亡,或程序性细胞死亡,通过消除受损或多余的细胞,在维持细胞稳态中起着关键作用。信号通路的失调,如JAK/STAT,与多种疾病有关,使它们成为潜在的新抗癌药物的有吸引力的治疗靶点。同时,保存TNF-α和p53等必需蛋白以维持正常的细胞生/死平衡是必要的。鉴于这些考虑,本研究采用了一种创新的硅混合和分层虚拟筛选方法,旨在确定JAK/STAT多靶点抑制剂作为几种肿瘤疾病的潜在抗癌药物。最初,生物靶标预测工具使用广泛的美国国家癌症研究所(NCI)数据库,以ON/ off靶标/多靶标组合模式使用,之前由ADME评估工具过滤。随后,我们对JAK2、JAK3和STAT3进行了分子对接研究,确定了最有希望的化合物755435。最后,分子动力学模拟验证了潜在的多靶点抑制剂755435与JAK2、JAK3和STAT3复合物的高稳定性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A novel in silico approach for identifying multi-target JAK/STAT inhibitors as anticancer agents
Apoptosis, or programmed cell death, plays a pivotal role in maintaining cellular homeostasis by eliminating damaged or surplus cells. Dysregulation of signaling pathways, such as JAK/STAT, is implicated in various diseases, rendering them attractive therapeutic targets for potential new anticancer drugs. Concurrently, it is imperative to preserve essential proteins like TNF-α and p53 to maintain normal cellular life/death balance. In light of these considerations, this study employs an innovative in silico hybrid and hierarchical virtual screening approach aimed at identifying JAK/STAT multi-target inhibitors as potential anticancer agents for several tumoral diseases. Initially, the Biotarget Predictor Tool is utilized in a combined ON/OFF-target/Multitarget mode using the extensive National Cancer Institute (NCI) database, previously filtered by ADME evaluation tools. Subsequently, Molecular Docking studies are conducted on JAK2, JAK3, and STAT3, facilitating the identification of the most promising compound, 755435. Finally, Molecular Dynamics Simulations validate the high stability of the potential multitarget inhibitor 755435 in complex with JAK2, JAK3, and STAT3.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


