基于深度学习和医学文本挖掘的临床试验资格标准的结构分析和智能分类。
IF 4
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
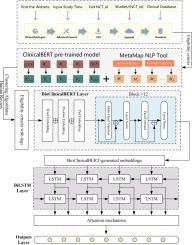
目的为了提高临床试验的效率、质量和创新能力,本文介绍了一种名为 CTEC-AC(临床试验资格标准自动分类)的新型模型,旨在将临床试验资格标准结构化为可计算解释的分类:方法:我们从 ClinicalTrials.gov 获取了最新 2,500 项临床试验的详细信息,生成了 20,000 多个资格标准数据条目。为了增强这些标准的表达能力,我们整合了两种强大的方法:ClinicalBERT 和 MetaMap。由此产生的增强特征被用作分层聚类算法的输入。后处理包括对算法输出的专家验证,以确保所构建的注释资格文本语料库的准确性。最终,我们的模型被用于对资格标准进行自动分类:结果:我们确定了 31 个不同的类别来概括临床研究人员撰写的资格标准,并发现了这些标准表达方式的共同主题。在标注数据集上使用我们的自动分类模型,我们取得了 0.94 的宏观平均 F1 分数:这项工作可以从非结构化的资格标准文本中自动提取结构化表述,极大地推动了临床试验的信息化进程。这反过来又能大大提高临床研究人员自动招募参与者的智能化程度。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structural analysis and intelligent classification of clinical trial eligibility criteria based on deep learning and medical text mining
Objective:
To enhance the efficiency, quality, and innovation capability of clinical trials, this paper introduces a novel model called CTEC-AC (Clinical Trial Eligibility Criteria Automatic Classification), aimed at structuring clinical trial eligibility criteria into computationally explainable classifications.
Methods:
We obtained detailed information on the latest 2,500 clinical trials from ClinicalTrials.gov, generating over 20,000 eligibility criteria data entries. To enhance the expressiveness of these criteria, we integrated two powerful methods: ClinicalBERT and MetaMap. The resulting enhanced features were used as input for a hierarchical clustering algorithm. Post-processing included expert validation of the algorithm’s output to ensure the accuracy of the constructed annotated eligibility text corpus. Ultimately, our model was employed to automate the classification of eligibility criteria.
Results:
We identified 31 distinct categories to summarize the eligibility criteria written by clinical researchers and uncovered common themes in how these criteria are expressed. Using our automated classification model on a labeled dataset, we achieved a macro-average F1 score of 0.94.
Conclusion:
This work can automatically extract structured representations from unstructured eligibility criteria text, significantly advancing the informatization of clinical trials. This, in turn, can significantly enhance the intelligence of automated participant recruitment for clinical researchers.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


