基于共识的深度半监督学习迭代元伪标记
IF 8.1
1区 计算机科学
0 COMPUTER SCIENCE, INFORMATION SYSTEMS
引用次数: 0
摘要
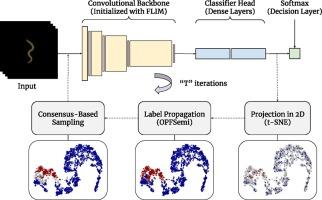
阻碍深度学习模型开发的一个已知问题是需要对大量样本进行准确标注,这是一项耗时、耗力且容易出错的任务。在数据标注需要专家知识的领域,这一限制尤为关键。伪标注等半监督学习方法可以利用有限的标注数据和大量的未标注数据来缓解这一问题;不过,最先进的方法通常需要预先训练编码器和验证集才能提供有效的解决方案。在此,我们介绍一种基于师生的迭代元伪标注方法,名为共识深度特征标注(consensus Deep Feature Annotation,简称cons-DeepFA),该方法可从少量标注样本中训练自定义卷积神经网络(CNN),而无需依赖预先训练的编码器和验证集、即用户在每类选定的几幅图像的判别区域上绘制标记。在每次迭代过程中,学生最后一个稠密层的潜在空间会通过一种基于最优连接性的方法(教师)非线性地投射到一个二维空间上,用于下游标签传播;之后,学生会使用由所提出的共识机制选择的伪标签样本进行再训练,随着迭代的进行,该机制会共同改善潜在空间、其投射以及学生的泛化能力。这种策略最近被引入到预训练编码器中,通过选择最有信心的伪标签样本来重新训练学生。Cons-DeepFA 以之前的方法为基础,做出了两大贡献。它(i)结合了 FLIM,从而能以更快的收敛速度从头开始训练自定义 CNN,提高其泛化能力;(ii)引入了基于共识的多次迭代程序,选择更准确的伪标签样本来重新训练 CNN。最后,cons-DeepFA 在五个具有挑战性的生物图像数据集上进行了评估,与来自四种半监督学习范式的七种最先进方法相比,证明了它的有效性和竞争力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Consensus-based iterative meta-pseudo-labeling for deep semi-supervised learning
A known issue that hinders the development of deep learning models is the need for accurate annotation of a large quantity of samples – a time-consuming, labor-intensive, and error-prone task. This limitation is particularly critical in areas where data annotation requires expert knowledge. Semi-supervised learning methods, such as pseudo-labeling, can alleviate the problem by capitalizing on both limited labeled and plentiful unlabeled data; nonetheless, state-of-the-art methods often require pre-trained encoders and validation sets to deliver effective solutions. Herein, we introduce a teacher-student-based iterative meta-pseudo-labeling approach, named consensus Deep Feature Annotation (cons-DeepFA), that enables the training of custom Convolutional Neural Networks (CNNs) from small quantities of labeled samples without reliance on pre-trained encoders and validation sets. cons-DeepFA explores Feature Learning from Image Markers (FLIM) to initialize the filters of a target CNN (student) from minimal data annotation – i.e., user-drawn markers on discriminative regions of a few selected images per class. During each of a few iterations, the latent space of the student's last dense layer is non-linearly projected onto a two-dimensional space for downstream label propagation via an optimum-connectivity-based approach (teacher); afterward, the student is re-trained using pseudo-labeled samples selected by the proposed consensus mechanism, which jointly improves the latent space, its projection, and the student's generalization ability as iterations progress. This strategy was recently introduced with pre-trained encoders by selecting the most confident pseudo-labeled samples to re-train the student. While building on previous methods, cons-DeepFA presents two key contributions. It (i) incorporates FLIM to enable training a custom CNN from scratch with faster convergence, improving its generalization ability, and (ii) introduces a consensus-based procedure over multiple iterations that selects more accurately pseudo-labeled samples for re-training the CNN. Lastly, cons-DeepFA is evaluated on five challenging biological image datasets, demonstrating its effectiveness and competitiveness when compared to seven state-of-the-art methods from four semi-supervised learning paradigms.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Information Sciences
工程技术-计算机:信息系统
CiteScore
14.00
自引率
17.30%
发文量
1322
审稿时长
10.4 months
期刊介绍:
Informatics and Computer Science Intelligent Systems Applications is an esteemed international journal that focuses on publishing original and creative research findings in the field of information sciences. We also feature a limited number of timely tutorial and surveying contributions.
Our journal aims to cater to a diverse audience, including researchers, developers, managers, strategic planners, graduate students, and anyone interested in staying up-to-date with cutting-edge research in information science, knowledge engineering, and intelligent systems. While readers are expected to share a common interest in information science, they come from varying backgrounds such as engineering, mathematics, statistics, physics, computer science, cell biology, molecular biology, management science, cognitive science, neurobiology, behavioral sciences, and biochemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


